

植物学报 ›› 2022, Vol. 57 ›› Issue (1): 42-55.DOI: 10.11983/CBB21158 cstr: 32102.14.CBB21158
王霞1, 严维1, 周志勤2, 常振仪1, 郑敏婷1, 唐晓艳1,2, 吴建新1,*( )
)
收稿日期:2021-09-14
接受日期:2021-11-17
出版日期:2022-01-01
发布日期:2022-01-17
通讯作者:
吴建新
作者简介:* E-mail: wjxin@m.scnu.edu.cn基金资助:
Xia Wang1, Wei Yan1, Zhiqin Zhou2, Zhenyi Chang1, Minting Zheng1, Xiaoyan Tang1,2, Jianxin Wu1,*( )
)
Received:2021-09-14
Accepted:2021-11-17
Online:2022-01-01
Published:2022-01-17
Contact:
Jianxin Wu
摘要: 水稻(Oryza sativa)隐性核雄性不育突变体是第三代杂交水稻技术的核心。为了挖掘优质雄性不育突变体, 该研究通过筛选优质籼稻黄华占(HHZ)的甲基磺酸乙酯(EMS)诱变突变体库, 获得1个雄性不育突变体ms102 (male sterility mutant 102)。该突变体营养生长正常, 但花药不开裂, 花粉败育。细胞学分析表明, 突变体花药绒毡层不能正常降解, 导致小孢子发育异常; 遗传分析表明, 该突变体的不育表型由1个已报道编码酰基转移酶的DPW2基因突变造成。研究获得了1个隐性核雄性不育突变体, 进一步证实了DPW2基因在水稻花药发育中的功能。
王霞, 严维, 周志勤, 常振仪, 郑敏婷, 唐晓艳, 吴建新. 水稻雄性不育突变体ms102的鉴定和基因定位. 植物学报, 2022, 57(1): 42-55.
Xia Wang, Wei Yan, Zhiqin Zhou, Zhenyi Chang, Minting Zheng, Xiaoyan Tang, Jianxin Wu. Identification and Mapping of a Rice Male Sterility Mutant ms102. Chinese Bulletin of Botany, 2022, 57(1): 42-55.
| Primers | Sequences (5′-3′) |
|---|---|
| OsACTIN-qPCR-F | GCTATGTACGTCGCCATCCA |
| OsACTIN-qPCR-R | GGACAGTGTGGCTGACACCAT |
| RTS-qPCR-F | ACATGTGGACTCGCTTGACT |
| RTS-qPCR-R | CATGGCTGCATGCAGATTCAT |
| TDR-qPCR-F | TGCTCTGGGAGCACAAGCC |
| TDR-qPCR-R | CTCGCTGTCCCTCACCATG |
| UDT1-qPCR-F | GAAGCACTCTGCAGCTAC |
| UDT1-qPCR-R | CTGCGTAGCCGTAGAAGG |
| DTM1-qPCR-F | AATTTAAGCCCTATCCCCTAAG |
| DTM1-qPCR-R | GAAGGTGGATTCCACTAGCTA |
| PTC1-qPCR-F | CACCAGATCATGGACCTCTG |
| PTC1-qPCR-R | AGCAGCCTCAGCTCCATGTG |
| DPW-qPCR-F | ACCACAGGAGACACGGTGATG |
| DPW-qPCR-R | CTTGGCCTGATGGTGACAA |
| PAIR1-qPCR-F | GGATGGACCCAGATTAACC |
| PAIR1-qPCR-R | CTGTTTAGGTGCCACCCTGT |
| PAIR2-qPCR-F | TGCCAGAGGAGAGGACCATTC |
| PAIR2-qPCR-R | CACGAGATGCTTGCTATTGAC |
| DTD-qPCR-F | TCATGAGTCTTTGAGCCGCA |
| DTD-qPCR-R | CACCTTCGCTCCAAATCTGT |
| OsCP1-qPCR-F | TGCAAGGATCACTCCACCTG |
| OsCP1-qPCR-R | GGCTGTTCTTGCTCTTGGAG |
| OsMSP1-qPCR-F | AGCATCGCGAGTAAGATGC |
| OsMSP1-qPCR-R | GTAGCCTTGTAACTTCAAGTAGGA |
| CYP703A3-qPCR-F | TGTACTGCTTTCTTTGCCCG |
| CYP703A3-qPCR-R | GCAGCAATCATGTCCTGCAT |
| CYP704B2-qPCR-F | GCTGGTTGATGACTTCACCT |
| CYP704B2-qPCR-R | CGACAGTATGTCGTGCTTGAT |
| OsGAMYB-qPCR-F | GCGACGGTATCATGTTCAAT |
| OsGAMYB-qPCR-R | GTCGCATAAGAGAACATCTG |
表1 qRT-PCR引物序列
Table 1 Sequences of primers for qRT-PCR
| Primers | Sequences (5′-3′) |
|---|---|
| OsACTIN-qPCR-F | GCTATGTACGTCGCCATCCA |
| OsACTIN-qPCR-R | GGACAGTGTGGCTGACACCAT |
| RTS-qPCR-F | ACATGTGGACTCGCTTGACT |
| RTS-qPCR-R | CATGGCTGCATGCAGATTCAT |
| TDR-qPCR-F | TGCTCTGGGAGCACAAGCC |
| TDR-qPCR-R | CTCGCTGTCCCTCACCATG |
| UDT1-qPCR-F | GAAGCACTCTGCAGCTAC |
| UDT1-qPCR-R | CTGCGTAGCCGTAGAAGG |
| DTM1-qPCR-F | AATTTAAGCCCTATCCCCTAAG |
| DTM1-qPCR-R | GAAGGTGGATTCCACTAGCTA |
| PTC1-qPCR-F | CACCAGATCATGGACCTCTG |
| PTC1-qPCR-R | AGCAGCCTCAGCTCCATGTG |
| DPW-qPCR-F | ACCACAGGAGACACGGTGATG |
| DPW-qPCR-R | CTTGGCCTGATGGTGACAA |
| PAIR1-qPCR-F | GGATGGACCCAGATTAACC |
| PAIR1-qPCR-R | CTGTTTAGGTGCCACCCTGT |
| PAIR2-qPCR-F | TGCCAGAGGAGAGGACCATTC |
| PAIR2-qPCR-R | CACGAGATGCTTGCTATTGAC |
| DTD-qPCR-F | TCATGAGTCTTTGAGCCGCA |
| DTD-qPCR-R | CACCTTCGCTCCAAATCTGT |
| OsCP1-qPCR-F | TGCAAGGATCACTCCACCTG |
| OsCP1-qPCR-R | GGCTGTTCTTGCTCTTGGAG |
| OsMSP1-qPCR-F | AGCATCGCGAGTAAGATGC |
| OsMSP1-qPCR-R | GTAGCCTTGTAACTTCAAGTAGGA |
| CYP703A3-qPCR-F | TGTACTGCTTTCTTTGCCCG |
| CYP703A3-qPCR-R | GCAGCAATCATGTCCTGCAT |
| CYP704B2-qPCR-F | GCTGGTTGATGACTTCACCT |
| CYP704B2-qPCR-R | CGACAGTATGTCGTGCTTGAT |
| OsGAMYB-qPCR-F | GCGACGGTATCATGTTCAAT |
| OsGAMYB-qPCR-R | GTCGCATAAGAGAACATCTG |
| Primers | Sequences (5'-3') |
|---|---|
| S1-HRM-F | GACAGGCAAGCAGAAAACCTG |
| S1-HRM-R | GTGCAGGTAGCACATCATCCT |
| S2-HRM-F | CTACAGATATATACAGGACGGTG |
| S2-HRM-R | GCTAAGGTAGCAACGTACACAG |
| S3-HRM-F | GATGGAGACTCACGTCCAGA |
| S3-HRM-R | CCACAGCCCTCATTTTCGT |
表2 高分辨率溶解曲线(HRM)引物序列
Table 2 Sequences of primers for high resolution melting (HRM)
| Primers | Sequences (5'-3') |
|---|---|
| S1-HRM-F | GACAGGCAAGCAGAAAACCTG |
| S1-HRM-R | GTGCAGGTAGCACATCATCCT |
| S2-HRM-F | CTACAGATATATACAGGACGGTG |
| S2-HRM-R | GCTAAGGTAGCAACGTACACAG |
| S3-HRM-F | GATGGAGACTCACGTCCAGA |
| S3-HRM-R | CCACAGCCCTCATTTTCGT |
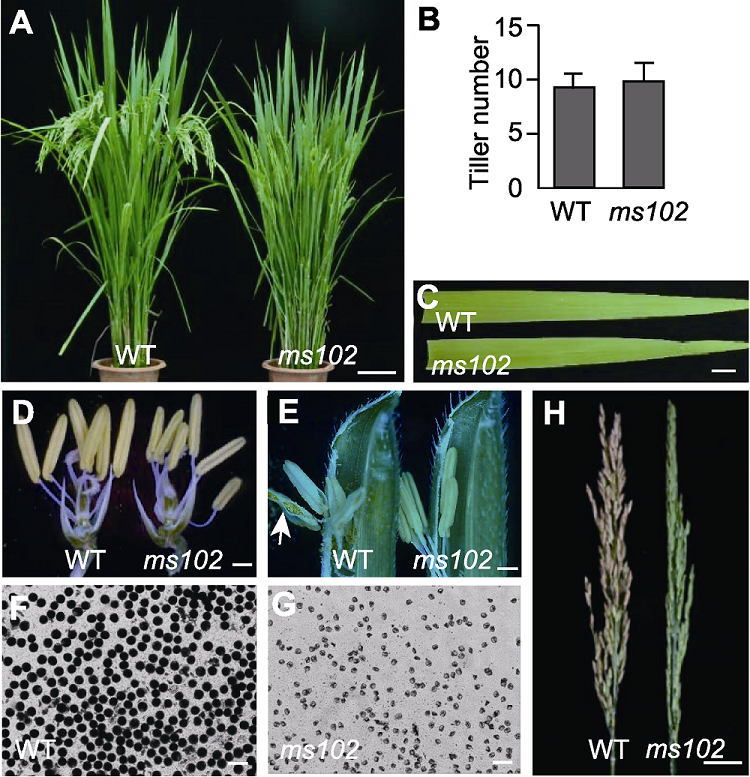
图1 水稻ms102突变体表型分析 (A) 成熟期植株的表型(bar=9 cm); (B) 成熟期野生型(WT)和ms102突变体分蘖数统计(n=23); (C) 扬花期野生型和ms102突变体叶色(bar=1 cm); (D) 去掉颖壳的成熟小花(bar=1 mm); (E) 扬花期的小花, 箭头指示开裂的花药(bar=1 cm); (F), (G) 野生型和ms102突变体的成熟花粉I2-KI染色(bars=50 μm); (H) 成熟期稻穗(bar=2.4 cm)
Figure 1 Phenotype analysis of the rice ms102 (A) Phenotype of mature plants (bar=9 cm); (B) Tiller number of mature wild type (WT) and ms102 plant (n=23); (C) Leaf color of WT and ms102 plant at flowering (bar=1 cm); (D) Mature spikelet without palea and lemma (bar=1 mm); (E) Flowering spikelet, the arrow indicates a dehisced anther (bar=1 cm); (F), (G) I2-KI-stained pollen of WT and ms102 mutant (bars=50 μm); (H) Mature panicles (bar=2.4 cm)
| Fertile plants | Male sterility plants | χ2(3:1) | χ20.05 |
|---|---|---|---|
| 359 | 102 | 2.034 | 3.841 |
表3 水稻WT × ms102杂交F2代表型分离比
Table 3 Segregation of the F2 progeny of rice WT × ms102
| Fertile plants | Male sterility plants | χ2(3:1) | χ20.05 |
|---|---|---|---|
| 359 | 102 | 2.034 | 3.841 |
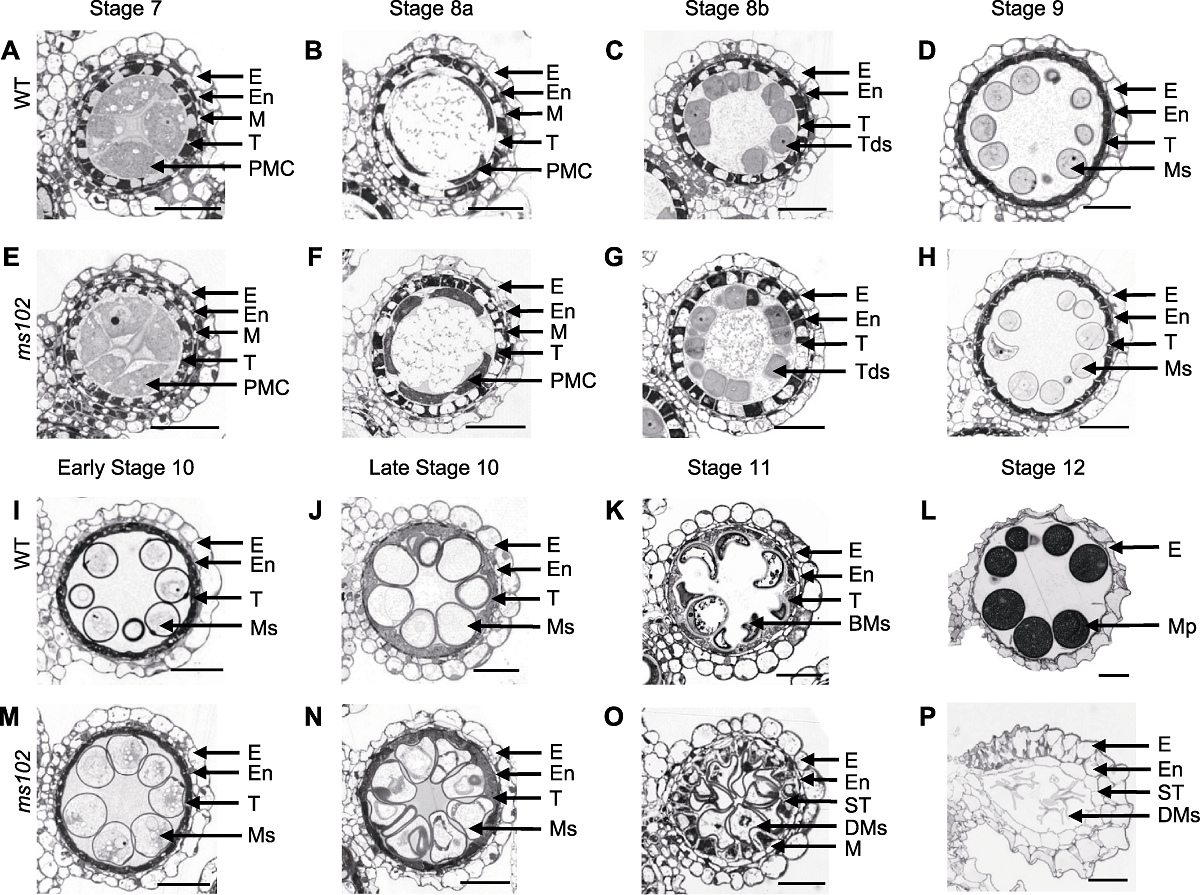
图2 水稻野生型(WT)和ms102突变体7-12时期花药切片 BMs: 二核小孢子; DMs: 降解小孢子; E: 外皮层; En: 内皮层; M: 中层; Mp: 成熟花粉; Ms: 小孢子; PMC: 花粉母细胞; ST: 膨大绒毡层; T: 绒毡层; Tds: 四分体。Bars=10 μm
Figure 2 Transverse sections images of WT and ms102 anthers at the developmental stages 7-12 BMs: Binuclear microspores; DMs: Degenerated microspores; E: Epidermis; En: Endothecium; M: Middle layer; Mp: Mature pollen; Ms: Microspores; PMC: Pollen mother cell; ST: Swollen tapetum; T: Tapetum; Tds: Tetrads. Bars=10 μm
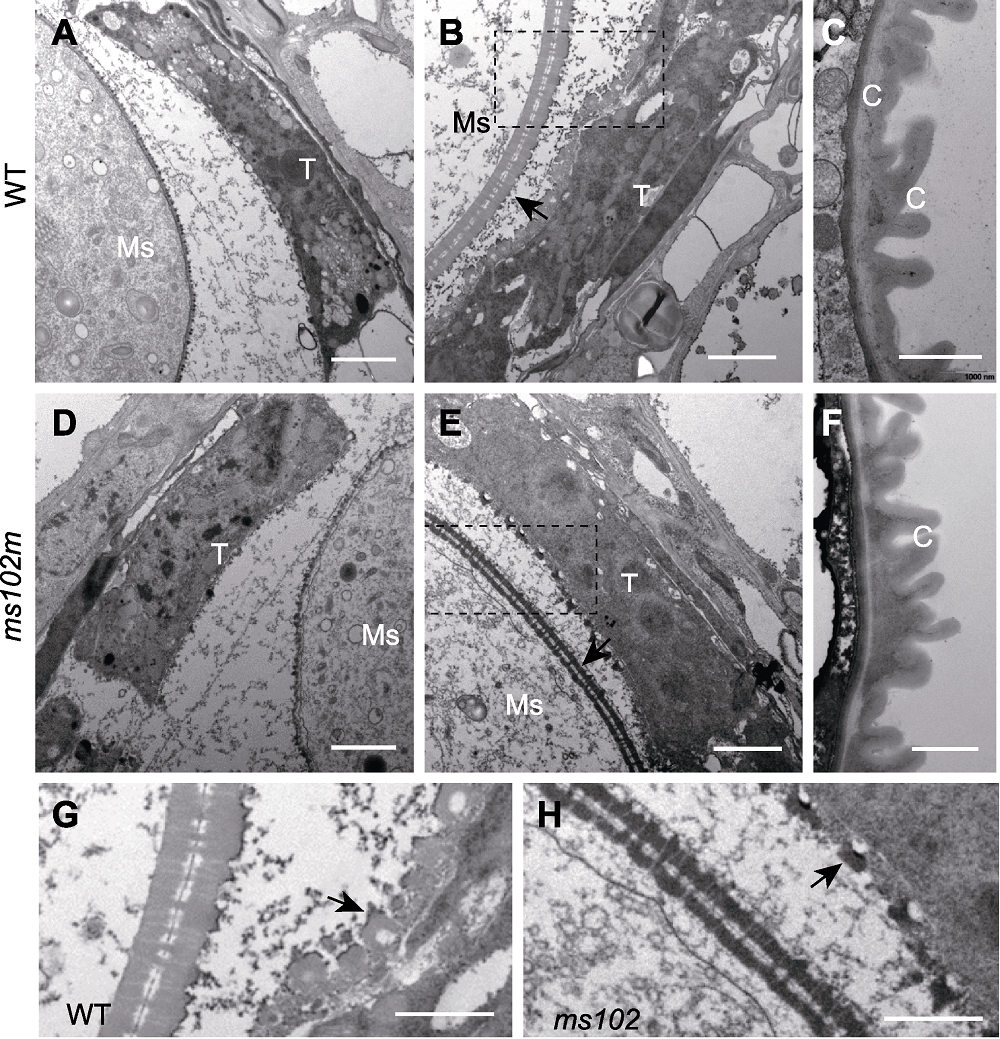
图3 水稻野生型(WT)和突变体(ms102)花药第9-12时期透射电镜超微结构 (A), (D) 第9时期; (B), (E) 第10时期(箭头指示小孢子外壁); (C), (F) 第12时期; (G), (H) 分别为B和E图中虚线框的放大部分(箭头指示乌氏体); T: 绒毡层; Ms: 小孢子; C: 角质层。(A), (B), (D), (E) Bars=2 μm; (C), (F), (G), (H) Bars =1 μm
Figure 3 Transmission electron microscope images of WT and ms102 anthers at the developmental stages 9-12 (A), (D) Stage 9; (B), (E) Stage 10 (the arrows indicate the microspore wall); (C), (F) Stage 12; (G), (H) The magnified region of dashed box in B and E (the arrows indicate the Ubisch bodies); T: Tapetum; Ms: Microspores; C: Cuticle. (A), (B), (D), (E) Bars=2 μm; (C), (F), (G), (H) Bars=1 μm
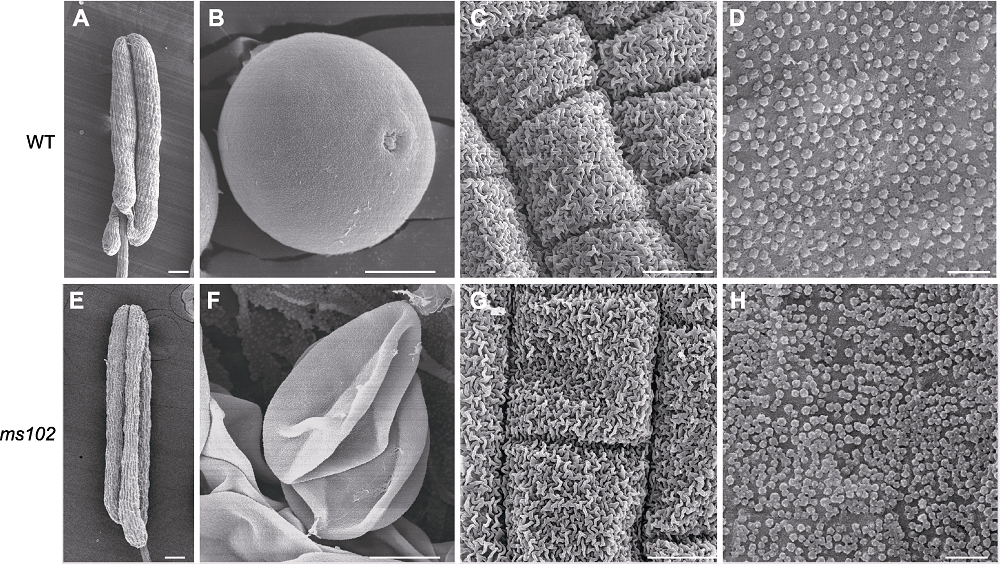
图4 水稻野生型(WT)和ms102突变体花药发育第12时期扫描电子显微镜超微结构 (A), (C), (E), (G) 花药外表面; (B), (F) 花粉粒; (D), (H) 乌氏体。(A), (E) Bars=100 μm; (B), (C), (F), (G) Bars=10 μm; (D), (H) Bars=2 μm
Figure 4 Scanning electron microscope images of WT and ms102 anthers at developmental stage 12 (A), (C), (E), (G) The external surfaces of anther wall; (B), (F) Pollen grains; (D), (H) Ubisch bodies. (A), (E) Bars =100 μm; (B), (C), (F), (G) Bars=10 μm; (D), (H) Bars=2 μm
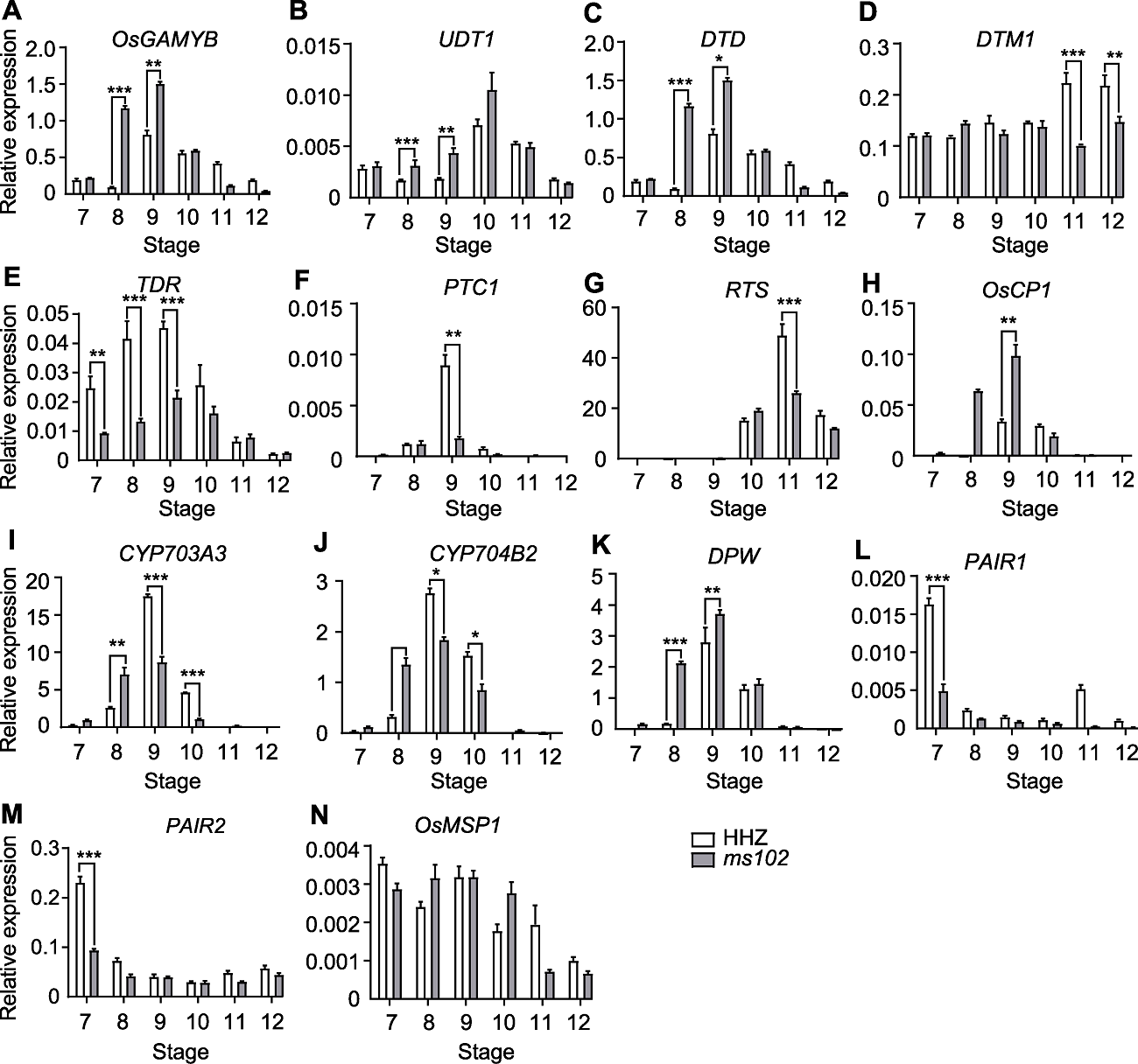
图5 花药发育相关基因在野生型和ms102花药中的表达 通过qRT-PCR检测野生型与ms102突变体中花药发育7-12时期相关基因的表达量。以OsACTIN1作为内参, 数据为平均值±标准差(n=3)。* P<0.05, ** P<0.01, *** P<0.001
Figure 5 Expression of genes related to pollen development in WT and ms102 anthers Anthers at stages 7-12 were used for qRT-PCR. OsACTIN1 served as an internal control. Data are shown as means±SD (n=3). * P<0.05, ** P<0.01, *** P<0.001
| Chr. locus | Physical location | Genotype | SNP index | Gene |
|---|---|---|---|---|
| Chr. 1 | 39,147,186 (S1) | C→T | 0.8966 | Intergenic |
| Chr. 1 | 39,167,971 (S2) | G→A | 0.8966 | Intergenic |
| Chr. 1 | 40,599,831 (S3) | C→T | 0.9474 | LOC_Os01g7-0140-LOC_O-s01g70150 |
表4 候选基因及注释信息
Table 4 Candidate genes and annotation information
| Chr. locus | Physical location | Genotype | SNP index | Gene |
|---|---|---|---|---|
| Chr. 1 | 39,147,186 (S1) | C→T | 0.8966 | Intergenic |
| Chr. 1 | 39,167,971 (S2) | G→A | 0.8966 | Intergenic |
| Chr. 1 | 40,599,831 (S3) | C→T | 0.9474 | LOC_Os01g7-0140-LOC_O-s01g70150 |
| The number of male sterile plants | The number of fertile plants | |||||
|---|---|---|---|---|---|---|
| Physical location | S1 | S2 | S3 | S1 | S2 | S3 |
| Wild type | 1 | 1 | 0 | 25 | 25 | 23 |
| Heterozygote | 8 | 8 | 0 | 19 | 19 | 21 |
| Mutant | 44 | 44 | 53 | 0 | 0 | 0 |
表5 3个SNP候选位点基因型-表型连锁分析
Table 5 Genotype-phenotype correlation analyses of the three candidate SNP sites
| The number of male sterile plants | The number of fertile plants | |||||
|---|---|---|---|---|---|---|
| Physical location | S1 | S2 | S3 | S1 | S2 | S3 |
| Wild type | 1 | 1 | 0 | 25 | 25 | 23 |
| Heterozygote | 8 | 8 | 0 | 19 | 19 | 21 |
| Mutant | 44 | 44 | 53 | 0 | 0 | 0 |
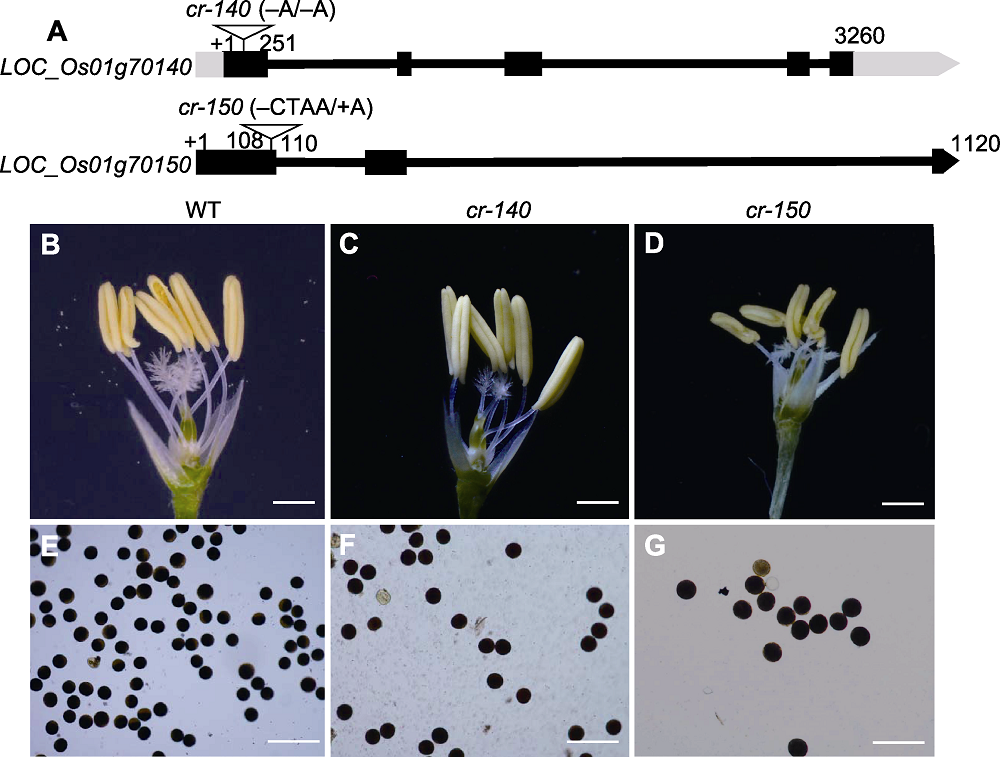
图6 水稻LOC_Os01g70140和LOC_Os01g70150基因结构及其突变体表型 (A) LOC_Os01g70140和LOC_Os01g70150基因结构及CRISPR突变体突变位点; (B)-(D) 花粉成熟期花药和雌蕊形态(bars=1 mm); (E)-(G) 成熟花粉粒I2-KI染色(bars=100 μm)。
Figure 6 Gene structures of LOC_Os01g70140 and LOC_Os01g70150 and mutant phenotypes (A) The gene structures and mutation sites of LOC_Os01g70140 and LOC_Os01g70150; (B)-(D) Morphology of mature anther and pistil (bars=1 mm); (E)-(G) Mature pollen grains stained with I2-KI (bars=100 μm).
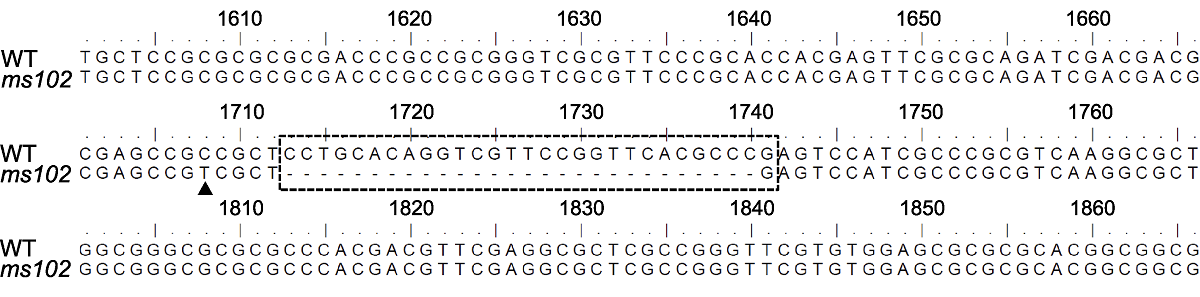
图7 水稻ms102突变体中DPW2基因突变位点分析 黑色三角形指示碱基替换突变位点, 虚线框指示缺失的核苷酸序列。
Figure 7 The mutation sites of DPW2 in ms102 The black arrow indicates the replaced nucleotide, the dashed box indicates the deleted nucleotides.
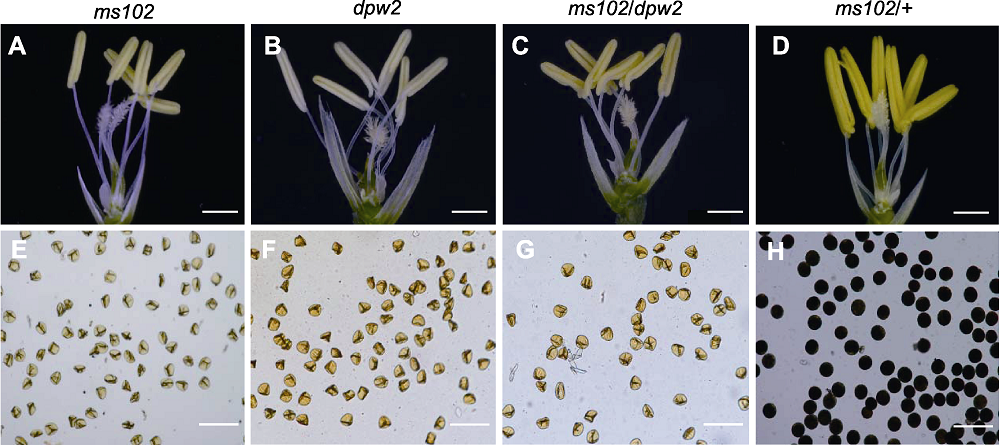
图8 水稻突变体dpw2/+ × ms102 F1的表型 (A)-(D) 花粉成熟期花药和雌蕊形态(bars=1 mm); (E)-(H) 成熟花粉粒I2-KI染色(bars=100 μm)。
Figure 8 The phenotype of the F1 progeny of dpw2/+ × ms102 The phenotype of the F1 progeny of dpw2/+ × ms102 (A)-(D) Morphology of mature anther and pistil (bars=1 mm); (E)-(H) Mature pollen grains stained with I2-KI (bars=100 μm).
| [1] | 陈竹锋, 卢嘉威, 卢启清, 王娜, 王成旭, 谢刚, 周向阳, 唐晓艳 (2014). 优质水稻黄华占雄性不育突变体的筛选及初步分析. 广东农业科学 41(19), 1-4. |
| [2] | 邓兴旺, 王海洋, 唐晓艳, 周君莉, 陈浩东, 何光明, 陈良碧, 许智宏 (2013). 杂交水稻育种将迎来新时代. 中国科学: 生命科学 43, 864-868. |
| [3] | 李文杰 (2019). 水稻花粉发育基因LSP2的克隆及功能研究. 硕士论文. 成都: 四川农业大学. pp. 1-2. |
| [4] | 倪浩凌, 吴文诗, 颜艳敏, 方亦圆, 王嘉茵, 陈碧湖, 李芷怡, 唐晓艳, 吴建新 (2020). 水稻穗发芽突变体的筛选及候选基因鉴定. 植物遗传资源学报 21, 1214-1220. |
| [5] |
张彤, 郭亚璐, 陈悦, 马金姣, 兰金苹, 燕高伟, 刘玉晴, 徐珊, 李莉云, 刘国振, 窦世娟 (2019). 水稻OsPR10A的表达特征及其在干旱胁迫应答过程中的功能. 植物学报 54, 711-722.
DOI |
| [6] | Abbas A, Yu P, Sun LP, Yang ZF, Chen DB, Cheng SH, Cao LY (2021). Exploiting genic male sterility in rice: from molecular dissection to breeding applications. Front Plant Sci 12, 629314. |
| [7] |
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30, 174-178.
DOI URL |
| [8] |
Ali Z, Raza Q, Atif RM, Aslam U, Ajmal M, Chung G (2019). Genetic and molecular control of floral organ iden- tity in cereals. Int J Mol Sci 20, 2743
DOI URL |
| [9] |
Ariizumi T, Toriyama K (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62, 437-460.
DOI PMID |
| [10] |
Bai WT, Wang PR, Hong J, Kong WY, Xiao YJ, Yu XW, Zheng H, You SM, Lu JY, Lei DK, Wang CL, Wang QM, Liu SJ, Liu X, Tian YL, Chen LM, Jiang L, Zhao ZG, Wu CY, Wan JM (2019). Earlier Degraded Tapetum 1 (EDT1) encodes an ATP-citrate lyase required for tapetum programmed cell death. Plant Physiol 181, 1223-1238.
DOI URL |
| [11] |
Bayer A, Ma XY, Stöckigt J (2004). Acetyltransfer in natural product biosynthesis-functional cloning and molecular analysis of vinorine synthase. Bioorg Med Chem 12, 2787-2795.
DOI URL |
| [12] |
Chang ZY, Chen ZF, Wang N, Xie G, Lu JW, Yan W, Zhou JL, Tang XY, Deng XW (2016a). Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci USA 113, 14145-14150.
DOI URL |
| [13] |
Chang ZY, Chen ZF, Yan W, Xie G, Lu JW, Wang N, Lu QQ, Yao N, Yang GZ, Xia JX, Tang XY (2016b). An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci 253, 21-30.
DOI URL |
| [14] |
Chang ZY, Jin MN, Yan W, Chen H, Qiu SJ, Fu S, Xia JX, Liu YC, Chen ZF, Wu JX, Tang XY (2018). The ATP- binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice 11, 58.
DOI URL |
| [15] |
Chang ZY, Xu CJ, Huang XY, Yan W, Qiu SJ, Yuan ST, Ni HL, Chen SJ, Xie G, Chen ZF, Wu JX, Tang XY (2020). The plant-specific ABERRANT GAMETOGENESIS 1 gene is essential for meiosis in rice. J Exp Bot 71, 204-218.
DOI URL |
| [16] |
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812-818.
DOI URL |
| [17] |
Gómez JF, Talle B, Wilson ZA (2015). Anther and pollen development: a conserved developmental pathway. J Integr Plant Biol 57, 876-891.
DOI URL |
| [18] |
Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, Comai L, Henikoff S (2003). Spectrum of chemically induced mutations from a large-scale reverse- genetic screen in Arabidopsis. Genetics 164, 731-740.
DOI URL |
| [19] |
Ji CH, Li HY, Chen LB, Xie M, Wang FP, Chen YL, Liu YG (2013). A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant 6, 1715-1718.
DOI URL |
| [20] |
Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005). Rice Undeveloped Tapetum 1 is a major regulator of early tapetum development. Plant Cell 17, 2705-2722.
PMID |
| [21] |
Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, Matsuoka M (2004). Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16, 33-44.
DOI URL |
| [22] | Kim YS, Schumaker KS, Zhu JK (2006). EMS mutagenesis of Arabidopsis. In: Salinas J, Sanchez-Serrano JJ, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 101- 103. |
| [23] |
Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao YW, Liang WQ, Zhang DB (2010a). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173-190.
DOI URL |
| [24] |
Li H, Yuan Z, Vizcay-Barrena G, Yang CY, Liang WQ, Zong J, Wilson ZA, Zhang DB (2011). PERSISTENT TAPETAL CELL 1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156, 615-630.
DOI URL |
| [25] |
Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, Luo D, Ma H, Zhang DB (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999-3014.
DOI URL |
| [26] |
Li YD, Chu ZZ, Liu XG, Jing HC, Liu YG, Hao DY (2010b). A cost-effective high-resolution melting approach using the EvaGreen dye for DNA polymorphism detection and genotyping in plants. J Integr Plant Biol 52, 1036-1042.
DOI URL |
| [27] |
Liao CC, Yan W, Chen ZF, Xie G, Deng XW, Tang XY (2021). Innovation and development of the third-generation hybrid rice technology. Crop J 9, 693-701.
DOI URL |
| [28] |
Liu ZH, Bao WJ, Liang WQ, Yin JY, Zhang DB (2010). Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J Integr Plant Biol 52, 670-678.
DOI URL |
| [29] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402-408.
DOI PMID |
| [30] |
Lochlainn SÓ, Amoah S, Graham NS, Alamer K, Rios JJ, Kurup S, Stoute A, Hammond JP, Østergaard L, King GJ, White PJ, Broadley MR (2011). High Resolution Melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods 7, 43.
DOI PMID |
| [31] |
Luo H, Lee JY, Hu Q, Nelson-Vasilchik K, Eitas TK, Lickwar C, Kausch AP, Chandlee JM, Hodges TK (2006). RTS, a rice anther-specific gene is required for male fertility and its promoter sequence directs tissue- specific gene expression in different plant species. Plant Mol Biol 62, 397-408.
DOI URL |
| [32] |
Ma H (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56, 393-434.
DOI URL |
| [33] |
Nonomura KI, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15, 1728-1739.
DOI URL |
| [34] |
Nonomura KI, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119, 217-225.
DOI URL |
| [35] |
Nonomura KI, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N (2004). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16, 1008-1020.
DOI URL |
| [36] |
Pan XY, Yan W, Chang ZY, Xu YC, Luo M, Xu CJ, Chen ZF, Wu JX, Tang XY (2020). OsMYB80 regulates anther development and pollen fertility by targeting multiple biological pathways. Plant Cell Physiol 61, 988-1004.
DOI URL |
| [37] |
Peng XQ, Wang ML, Li YQ, Yan W, Chang ZY, Chen ZF, Xu CJ, Yang CW, Deng XW, Wu JX, Tang XY (2020). Lectin receptor kinase OsLecRK-S.7 is required for pollen development and male fertility. J Integr Plant Biol 62, 1227-1245.
DOI URL |
| [38] |
Quilichini TD, Grienenberger E, Douglas CJ (2015). The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113, 170-182.
DOI PMID |
| [39] |
Shi J, Tan HX, Yu XH, Liu YY, Liang WQ, Ranathunge K, Franke RB, Schreiber L, Wang YJ, Kai GY, Shanklin J, Ma H, Zhang DB (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23, 2225-2246.
DOI URL |
| [40] |
Suzuki H, Nakayama T, Nishino T (2003). Proposed mecha- nism and functional amino acid residues of malonyl-CoA: anthocyanin 5-O-glucoside-6'''-O-malonyltransferase from flowers of Salvia splendens, a member of the versatile plant acyltransferase family. Biochemistry 42, 1764-1771.
DOI URL |
| [41] |
Wang B, Fang RQ, Chen FM, Han JL, Liu YG, Chen LT, Zhu QL (2020). A novel CCCH-type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice. J Integr Plant Biol 62, 1594-1606.
DOI URL |
| [42] | Xu DW, Shi JX, Rautengarten C, Yang L, Qian XL, Uzair M, Zhu L, Luo Q, An G, Waßmann F, Schreiber L, Heazlewood JL, Scheller HV, Hu JP, Zhang DB, Liang WQ (2017). DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol 173, 240-255. |
| [43] | Yan W, Chen ZF, Lu JW, Xu CJ, Xie G, Li YQ, Deng XW, He H, Tang XY (2017). Simultaneous identification of multiple causal mutations in rice. Front Plant Sci 7, 2055. |
| [44] |
Yan W, Deng XW, Yang C, Tang X (2021). The genome-wide EMS mutagenesis bias correlates with sequence context and chromatin structure in rice. Front Plant Sci 12, 579675.
DOI URL |
| [45] |
Yang XJ, Wu D, Shi JX, He Y, Pinot F, Grausem B, Yin CS, Zhu L, Chen MJ, Luo ZJ, Liang W, Zhang D (2014). Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56, 979-994.
DOI URL |
| [46] |
Yang ZF, Liu L, Sun LP, Yu P, Zhang PP, Abbas A, Xiang XJ, Wu WX, Zhang YX, Cao LY, Cheng SH (2019). OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol Biol 99, 175-191.
DOI URL |
| [47] |
Yi J, Kim SR, Lee DY, Moon S, Lee YS, Jung KH, Hwang I, An G (2012). The rice gene DEFECTIVE TAPETUM AND MEIOCYTES 1 (DTM1) is required for early tapetum development and meiosis. Plant J 70, 256-270.
DOI URL |
| [48] | Zhang DB, Luo X, Zhu L (2011). Cytological analysis and genetic control of rice anther development. J Genet Genomics 38, 379-390. |
| [49] |
Zhang HH, Wang ML, Li YQ, Yan W, Chang ZY, Ni HL, Chen ZF, Wu JX, Xu CJ, Deng XW, Tang XY (2020). GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 are essential for rice pollen development. J Integr Plant Biol 62, 1574-1593.
DOI URL |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 王子阳, 刘升学, 杨志蕊, 秦峰. 玉米抗旱性的遗传解析[J]. 植物学报, 2024, 59(6): 883-902. |
| [6] | 吴锁伟, 安学丽, 万向元. 玉米雄性不育机理及其在工程核不育制种中的应用[J]. 植物学报, 2024, 59(6): 932-949. |
| [7] | 郑名敏, 黄强, 张鹏, 刘孝伟, 赵卓凡, 易洪杨, 荣廷昭, 曹墨菊. 玉米细胞质雄性不育及育性恢复研究进展[J]. 植物学报, 2024, 59(6): 999-1006. |
| [8] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [9] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [10] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [11] | 何璐梅, 马伯军, 陈析丰. 植物执行者抗病基因研究进展[J]. 植物学报, 2024, 59(4): 671-680. |
| [12] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [13] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [14] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [15] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||