

植物学报 ›› 2025, Vol. 60 ›› Issue (3): 449-459.DOI: 10.11983/CBB24165 cstr: 32102.14.CBB24165
收稿日期:2024-10-30
接受日期:2025-01-20
出版日期:2025-05-10
发布日期:2025-01-21
通讯作者:
*韩榕, 二级教授, 博士生导师。长期从事植物逆境响应研究, 重点关注UV-B辐射下小麦和拟南芥等植物在分子细胞学水平上的响应机制。主持完成多项国家自然科学基金和山西省自然科学基金项目。在Plant Physiology and Biochemistry、Environmental Science: Nano及Cell Research等国际重要核心期刊上发表高水平研究论文100余篇。曾获“山西省高等学校科技进步奖”一等奖和二等奖以及“陕西省科学技术奖”二等奖等奖项。E-mail: hanrong@sxnu.edu.cn
基金资助:
Chen Pengxiang, Wang Bo, Wang Zijun, Han Rong*( )
)
Received:2024-10-30
Accepted:2025-01-20
Online:2025-05-10
Published:2025-01-21
Contact:
*E-mail: hanrong@sxnu.edu.cn
摘要: 作为太阳光的固有成分, UV-B对植物生长发育有重要影响。随着对UV-B研究的深入, 人们认识到UV-B不仅是环境胁迫因子, 还是植物生长过程中的重要信号分子, 适度的UV-B辐射对植物生长具有促进作用。UVR8是UV-B特有的光感受器, 在植物响应UV-B过程中发挥不可替代的作用, 且其功能受上、下游转录因子调节。目前, 已知BBX、WRKY、MYB和PIF等多种转录因子参与调控UV-B辐射下的下胚轴伸长、主根长度、叶片大小及形态、开花周期和花青素合成等过程。该文主要综述了UVR8在UV-B信号通路中的分子机制, 并对转录因子在UV-B辐射过程中的调控机理进行总结, 以期为相关研究提供参考。
陈鹏翔, 王波, 王子俊, 韩榕. 转录因子在植物响应UV-B辐射中的调控作用. 植物学报, 2025, 60(3): 449-459.
Chen Pengxiang, Wang Bo, Wang Zijun, Han Rong. The Regulatory Roles of the Transcription Factors in Plant's Response to UV-B Radiation. Chinese Bulletin of Botany, 2025, 60(3): 449-459.
| 家族 | 成员 | 顺式元件 | 作用 | 参考文献 |
|---|---|---|---|---|
| bZIP | HY5 | T/G-box、E-box、GATA-box、ACE- box、Z-box和C-box | 调节下胚轴伸长, 在植物色素合成和光形态建成中起核心作用 | Stracke et al., |
| WRKY | WRKY36 | W-box | 促进下胚轴伸长及叶片发育 | Yang et al., |
| WRKY32 | W-box | 促进下胚轴伸长, 抑制拟南芥子叶展开 | Zhou et al., | |
| MYB | MYB11、MYB12和MYB111 | R2R3-MYB-binding motif | 抑制下胚轴伸长, 调控花青素生物合成, 影响次生代谢物合成 | Stracke et al., |
| MYB73/77 | R2R3-MYB-binding motif | 正调控生长素响应基因的表达, 促进下胚轴伸长和侧根生长 | Yang et al., | |
| BBX | BBX11 | T/G-BOX | 调节拟南芥下胚轴伸长, 参与植物光形态建成 | Job et al., |
| BBX20、BBX21、BBX22和BBX31 | T/G-BOX | 光形态建成正调控因子 | Heng et al., | |
| BBX24 | T/G-BOX | 光形态建成负调控因子 | Jiang et al., | |
| PIF | PIF4和PIF5 | G-box | 抑制下胚轴伸长, 参与植物光形态建成 | Sharma et al., |
| TCP | TCP4 | GGACCAC | 调控植物生长发育, 参与光响应 | Li et al., |
| BES | BES1/BIM1 | E-box和CGTGT/CG | 调节植物激素信号转导, 影响植物生长发育 | Liang et al., |
表1 转录因子在植物响应UV-B信号通路中的作用
Table 1 Function of transcription factors in plant response UV-B radiation
| 家族 | 成员 | 顺式元件 | 作用 | 参考文献 |
|---|---|---|---|---|
| bZIP | HY5 | T/G-box、E-box、GATA-box、ACE- box、Z-box和C-box | 调节下胚轴伸长, 在植物色素合成和光形态建成中起核心作用 | Stracke et al., |
| WRKY | WRKY36 | W-box | 促进下胚轴伸长及叶片发育 | Yang et al., |
| WRKY32 | W-box | 促进下胚轴伸长, 抑制拟南芥子叶展开 | Zhou et al., | |
| MYB | MYB11、MYB12和MYB111 | R2R3-MYB-binding motif | 抑制下胚轴伸长, 调控花青素生物合成, 影响次生代谢物合成 | Stracke et al., |
| MYB73/77 | R2R3-MYB-binding motif | 正调控生长素响应基因的表达, 促进下胚轴伸长和侧根生长 | Yang et al., | |
| BBX | BBX11 | T/G-BOX | 调节拟南芥下胚轴伸长, 参与植物光形态建成 | Job et al., |
| BBX20、BBX21、BBX22和BBX31 | T/G-BOX | 光形态建成正调控因子 | Heng et al., | |
| BBX24 | T/G-BOX | 光形态建成负调控因子 | Jiang et al., | |
| PIF | PIF4和PIF5 | G-box | 抑制下胚轴伸长, 参与植物光形态建成 | Sharma et al., |
| TCP | TCP4 | GGACCAC | 调控植物生长发育, 参与光响应 | Li et al., |
| BES | BES1/BIM1 | E-box和CGTGT/CG | 调节植物激素信号转导, 影响植物生长发育 | Liang et al., |
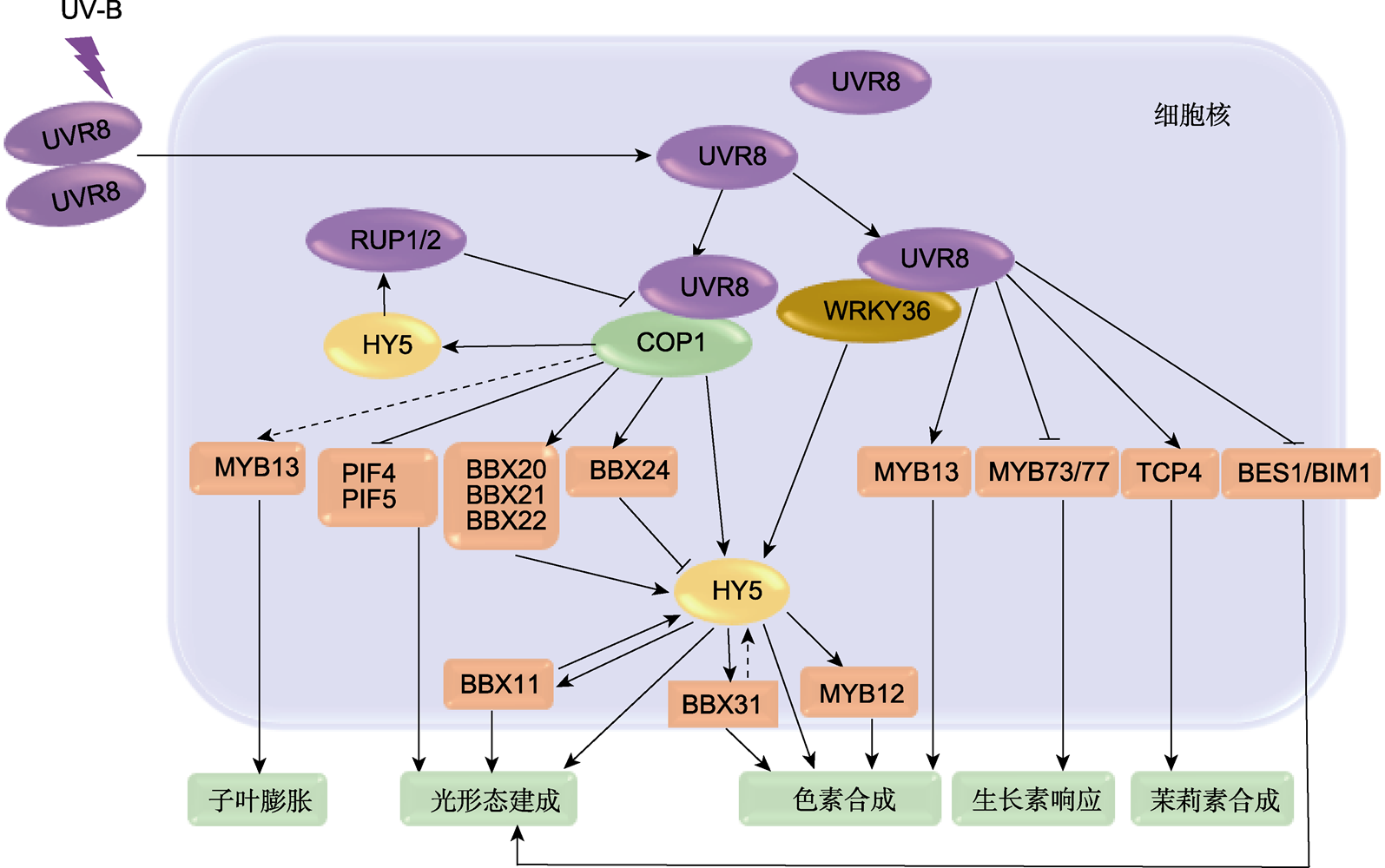
图1 转录因子参与植物UV-B信号通路模型 紫外线B (UV-B)激活其受体UVR8后, UVR8解聚为单体进入细胞核, 进而引发下游信号通路。UVR8直接与E3泛素连接酶COP1作用, 释放COP1降解底物HY5以及BBX20-22、BBX24、PIF4和PIF5转录因子。同时, UVR8与转录因子WRKY36相互作用增强和稳定HY5的转录活性, HY5启动下游转录因子BBX11、BBX31和MYB12, 从而调控植物色素合成与光形态建成。UVR8可抑制MYB73/MYB77和BES1/BIM1的DNA结合活性, 也可促进MYB13和TCP4对下游基因的激活, 从而调控下游信号通路。实线箭头和实线T型分别表示激活和抑制; 虚线表示有待实验验证。
Figure 1 Model of transcription factors involved in plant's UV-B signaling pathway Upon ultraviolet-B (UV-B) irradiation, its receptor UVR8 gets activated and subsequently depolymerizes into monomers, which then are translocated into the nucleus, instigating the downstream signaling pathways. UVR8 directly interacts with the E3 ubiquitin ligase COP1, resulting in the release of HY5, as well as the transcription factors BBX20-22, 24, PIF4 and PIF5 that would otherwise be degraded by COP1. On the other hand, UVR8 interacts with the transcription factor WRKY36 to potentiate and stabilize the transcriptional activity of HY5. HY5 triggers the downstream transcription factors BBX11, BBX31 and MYB12, orchestrating the regulation of plant pigment biosynthesis and photomorphogenesis. Moreover, UVR8 can inhibit the DNA-binding activities of MYB73/MYB77 and BES1/BIM1, and also promote the activation of downstream gene initiation by MYB13 and TCP4, thereby regulating the downstream signaling pathways. Solid arrows and T-lines represent activation and repression, respectively; the dotted lines represent that to be verified.
| [1] |
Bai SL, Saito T, Honda C, Hatsuyama Y, Ito A, Moriguchi T (2014). An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240, 1051-1062.
DOI PMID |
| [2] | Binkert M, Kozma-Bognár L, Terecskei K, de Veylder L, Nagy F, Ulm R (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200-4213. |
| [3] |
Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102, 18225-18230.
DOI PMID |
| [4] | Brown BA, Headland LR, Jenkins GI (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem Photobiol 85, 1147-1155. |
| [5] |
Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H (2020). Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 6, 921-928.
DOI PMID |
| [6] | Cai YP, Liu YT, Fan YY, Li XT, Yang MS, Xu DQ, Wang HY, Deng XW, Li J (2023). MYB112 connects light and circadian clock signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 35, 3485-3503. |
| [7] | Cao YP, Li K, Li YL, Zhao XP, Wang LH (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biology 9, 61. |
| [8] | Chen HZ, Han R (2015). Plants respond to UV-B radiation: a review. Chin Bull Bot 50, 790-801. (in Chinese) |
|
陈慧泽, 韩榕 (2015). 植物响应UV-B辐射的研究进展. 植物学报 50, 790-801.
DOI |
|
| [9] | Chen HZ, Niu JR, Han R (2021). Signal transduction pathways of plant ultraviolet B receptor UVR8. Plant Physiol J 57, 1179-1188. (in Chinese) |
| 陈慧泽, 牛靖蓉, 韩榕 (2021). 植物紫外光B受体UVR8的信号转导途径. 植物生理学报 57, 1179-1188. | |
| [10] | Chen S, Podolec R, Arongaus AB, Fuchs C, Loubéry S, Demarsy E, Ulm R (2024). Functional divergence of Arabidopsis REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 and 2 in repression of flowering. Plant Physiol 194, 1563-1576. |
| [11] | Crestani G, Cunningham N, Csepregi K, Badmus UO, Jansen MAK (2023). From stressor to protector, UV-induced abiotic stress resistance. Photochem Photobiol Sci 22, 2189-2204. |
| [12] | Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324-2338. |
| [13] |
Demarsy E (2022). WRKY-ing in the light. New Phytol 235, 5-7.
DOI PMID |
| [14] | Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci 15, 573-581. |
| [15] |
Fang F, Lin L, Zhang QW, Lu M, Skvortsova MY, Podolec R, Zhang QY, Pi JH, Zhang CL, Ulm R, Yin RH (2022). Mechanisms of UV-B light-induced photoreceptor UVR8 nuclear localization dynamics. New Phytol 236, 1824-1837.
DOI PMID |
| [16] | Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, Seidlitz HK, Nagy F, Ulm R (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28, 591-601. |
| [17] | Gaillochet C, Burko Y, Platre MP, Zhang L, Simura J, Willige BC, Kumar SV, Ljung K, Chory J, Busch W (2020). HY5 and phytochrome activity modulate shoot-to-root coordination during thermomorphogenesis in Arabidopsis. Development 147, dev192625. |
| [18] |
Gangappa SN, Botto JF (2014). The BBX family of plant transcription factors. Trends Plant Sci 19, 460-470.
DOI PMID |
| [19] |
Gangappa SN, Botto JF (2016). The multifaceted roles of HY5 in plant growth and development. Mol Plant 9, 1353-1365.
DOI PMID |
| [20] | Goyal P, Devi R, Verma B, Hussain S, Arora P, Tabassum R, Gupta S (2023). WRKY transcription factors: evolution, regulation, and functional diversity in plants. Protoplasma 260, 331-348. |
| [21] | Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107, 20132-20137. |
| [22] |
Guilfoyle TJ, Hagen G (2007). Auxin response factors. Curr Opin Plant Biol 10, 453-460.
DOI PMID |
| [23] | Han X, Huang X, Deng XW (2020). The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun 1, 100044. |
| [24] |
Haves S, Sharma A, Fraser DP, Trevisan M, Cragg-Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA (2017). UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol 27, 120-127.
DOI PMID |
| [25] | Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014). UV- B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA 111, 11894-11899. |
| [26] | Heijde M, Ulm R (2012). Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA 110, 1113-1118. |
| [27] | Heng YQ, Lin F, Jiang Y, Ding MQ, Yan TT, Lan HX, Zhou H, Zhao XH, Xu DQ, Deng XW (2019). B-box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol 180, 497-508. |
| [28] | Huang X, Ouyang XH, Yang PY, Lau OS, Li G, Li JG, Chen HD, Deng XW (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24, 4590-4606. |
| [29] |
Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS (2012). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22, 1046-1057.
DOI PMID |
| [30] |
Job N, Lingwan M, Masakapalli SK, Datta S (2022). Transcription factors BBX11 and HY5 interdependently regulate the molecular and metabolic responses to UV-B. Plant Physiol 189, 2467-2480.
DOI PMID |
| [31] | Kim BC, Tennessen DJ, Last RL (1998). UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 15, 667-674. |
| [32] | Küpers JJ, Oskam L, Pierik R (2020). Photoreceptors regulate plant developmental plasticity through auxin. Plants 9, 940. |
| [33] | Li C, Du JC, Xu HN, Feng ZH, Chater CCC, Duan YW, Yang YP, Sun XD (2024). UVR8-TCP4-LOX2 module regulates UV-B tolerance in Arabidopsis. J Integr Plant Biol 66, 897-908. |
| [34] | Li CX, Yu WJ, Xu JR, Lu XF, Liu YZ (2022). Anthocyanin biosynthesis induced by MYB transcription factors in plants. Int J Mol Sci 23, 11701. |
| [35] |
Li XL, Ren QL, Zhao WX, Liao CC, Wang Q, Ding TH, Hu H, Wang M (2023). Interaction between UV-B and plant anthocyanins. Funct Plant Biol 50, 599-611.
DOI PMID |
| [36] | Liang T, Mei SL, Shi C, Yang Y, Peng Y, Ma LB, Wang F, Li X, Huang X, Yin YH, Liu HT (2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44, 512-523. |
| [37] | Liang T, Shi C, Peng Y, Tan HJ, Xin PY, Yang Y, Wang F, Li X, Chu JF, Huang JR, Yin YH, Liu HT (2020). Brassinosteroid-activated BRI1-EMS-SUPPRESSOR 1 inhibits flavonoid biosynthesis and coordinates growth and UV-B stress responses in plants. Plant Cell 32, 3224-3239. |
| [38] |
Liang T, Yang Y, Liu HT (2019). Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol 221, 1247-1252.
DOI PMID |
| [39] | Lin N, Liu XY, Zhu WF, Cheng X, Wang XH, Wan XC, Liu LL (2021). Ambient ultraviolet B signal modulates tea flavor characteristics via shifting a metabolic flux in flavonoid biosynthesis. J Agric Food Chem 69, 3401-3414. |
| [40] | Liu MX, Sun M, Wang Y, Li YH (2012). Arabidopsis UV-B photoreceptor and its light signal transduction in plants. Chin Bull Bot 47, 661-669. (in Chinese) |
|
刘明雪, 孙梅, 王宇, 李玉花 (2012). 植物UV-B受体及其介导的光信号转导. 植物学报 47, 661-669.
DOI |
|
| [41] |
Lockhart J (2014). How ELONGATED HYPOCOTYL5 helps protect plants from UV-B rays. Plant Cell 26, 3826.
DOI |
| [42] | Lyu GZ, Li DB, Li SS (2020). Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal Behav 15, 1782647. |
| [43] | Miao TT, Li DZ, Huang ZY, Huang YW, Li SS, Wang Y (2021). Gibberellin regulates UV-B-induced hypocotyl growth inhibition in Arabidopsis thaliana. Plant Signal Behav 16, 1966587. |
| [44] | Mosadegh H, Trivellini A, Lucchesini M, Ferrante A, Maggini R, Vernieri P, Mensuali Sodi A (2019). UV-B physiological changes under conditions of distress and eustress in sweet basil. Plants 8, 396. |
| [45] | Ng DWK, Abeysinghe JK, Kamali M (2018). Regulating the regulators: the control of transcription factors in plant defense signaling. Int J Mol Sci 19, 3737. |
| [46] |
Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Ádám É, Schäfer E, Nagy F, Ulm R (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975-1990.
DOI PMID |
| [47] |
Petroni K, Falasca G, Calvenzani V, Allegra D, Stolfi C, Fabrizi L, Altamura MM, Tonelli C (2008). The AtMYB11 gene from Arabidopsis is expressed in meristematic cells and modulates growth in planta and organogenesis in vitro. J Exp Bot 59, 1201-1213.
DOI PMID |
| [48] |
Podolec R, Demarsy E, Ulm R (2021). Perception and signaling of ultraviolet-B radiation in plants. Annu Rev Plant Biol 72, 793-822.
DOI PMID |
| [49] |
Podolec R, Ulm R (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol 45, 18-25.
DOI PMID |
| [50] | Podolec R, Wagnon TB, Leonardelli M, Johansson H, Ulm R (2022). Arabidopsis B-box transcription factors BBX20- 22 promote UVR8 photoreceptor-mediated UV-B responses. Plant J 111, 422-439. |
| [51] | Ponnu J (2022). B-BOXing against UV rays: the BBX11- HY5 feedback loop regulates plant ultraviolet B tolerance. Plant Physiol 189, 1904-1905. |
| [52] |
Qian CZ, Chen ZR, Liu Q, Mao WW, Chen YL, Tian W, Liu Y, Han JP, Ouyang XH, Huang X (2020). Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses. Mol Plant 13, 777-792.
DOI PMID |
| [53] | Qian CZ, Mao WW, Liu Y, Ren H, Lau OS, Ouyang XH, Huang X (2016). Dual-source nuclear monomers of UV-B light receptor direct photomorphogenesis in Arabidopsis. Mol Plant 9, 1671-1674. |
| [54] |
Rai N, Neugart S, Yan Y, Wang F, Siipola SM, Lindfors AV, Winkler JB, Albert A, Brosché M, Lehto T, Morales LO, Aphalo PJ (2019). How do cryptochromes and UVR8 interact in natural and simulated sunlight? J Exp Bot 70, 4975-4990.
DOI PMID |
| [55] | Ratcliffe OJ, Riechmann JL (2002). Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective. Curr Issues Mol Biol 4, 77-91. |
| [56] | Saini P, Bhatia S, Mahajan M, Kaushik A, Sahu SK, Kumar A, Satbhai SB, Patel MK, Saxena S, Chaurasia OP, Lingwan M, Masakapalli SK, Yadav RK (2020). ELONGATED HYPOCOTYL5 negatively regulates DECREASE WAX BIOSYNTHESIS to increase survival during UV-B stress. Plant Physiol 184, 2091-2106. |
| [57] | Sharma A, Pridgeon AJ, Liu W, Segers F, Sharma B, Jenkins GI, Franklin KA (2023). ELONGATED HYPOCOTYL5 (HY5) and HY5 HOMOLOGUE (HYH) maintain shade avoidance suppression in UV-B. Plant J 115, 1394-1407. |
| [58] |
Sharma A, Sharma B, Hayes S, Kerner K, Hoecker U, Jenkins GI, Franklin KA (2019). UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat Commun 10, 4417.
DOI PMID |
| [59] |
Shi C, Liu HT (2021). How plants protect themselves from ultraviolet-B radiation stress. Plant Physiol 187, 1096-1103.
DOI PMID |
| [60] | Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/ PAP1 transcription factor in Arabidopsis. FEBS Lett 587, 1543-1547. |
| [61] | Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010a). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33, 88-103. |
| [62] |
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50, 660-677.
DOI PMID |
| [63] | Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010b). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188, 985-1000. |
| [64] | Strader L, Weijers D, Wagner D (2022). Plant transcription factors—being in the right place with the right company. Curr Opin Plant Biol 65, 102136. |
| [65] | Tavridou E, Schmid-Siegert E, Fankhauser C, Ulm R (2020). UVR8-mediated inhibition of shade avoidance involves HFR1 stabilization in Arabidopsis. PLoS Genet 16, e1008797. |
| [66] | Verma S, Attuluri VPS, Robert HS (2022). Transcriptional control of Arabidopsis seed development. Planta 255, 90. |
| [67] | Wang LX, Wang YD, Chang HF, Ren H, Wu XQ, Wen J, Guan ZY, Ma L, Qiu L, Yan JJ, Zhang DL, Huang X, Yin P (2023a). RUP2 facilitates UVR8 redimerization via two interfaces. Plant Commun 4, 100428. |
| [68] | Wang WL, Paik I, Kim J, Hou XL, Sung S, Huq E (2021a). Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis. New Phytol 230, 2311-2326. |
| [69] | Wang XP, Niu YL, Zheng Y (2021b). Multiple functions of MYB transcription factors in abiotic stress responses. Int J Mol Sci 22, 6125. |
| [70] | Wang YD, Wang LX, Guan ZY, Chang HF, Ma L, Shen CC, Qiu L, Yan JJ, Zhang DL, Li J, Deng XW, Yin P (2022). Structural insight into UV-B-activated UVR8 bound to COP1. Sci Adv 8, eabn3337. |
| [71] | Wang YJ, Zhou HY, He YR, Shen XP, Lin S, Huang L (2023b). MYB transcription factors and their roles in the male reproductive development of flowering plants. Plant Sci 335, 111811. |
| [72] | Weidemüller P, Kholmatov M, Petsalaki E, Zaugg JB (2021). Transcription factors: bridge between cell signaling and gene regulation. Proteomics 21, 2000034. |
| [73] | Wu XJ, Chen BC, Xiao JP, Guo HC (2023). Different doses of UV-B radiation affect pigmented potatoes' growth and quality during the whole growth period. Front Plant Sci 14, 1101172. |
| [74] | Xu DQ (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228, 1748-1753. |
| [75] |
Xu Y, Zhu ZQ (2020). UV-B response: when UVR8 meets MYBs. Trends Plant Sci 25, 515-517.
DOI PMID |
| [76] | Yadav A, Bakshi S, Yadukrishnan P, Lingwan M, Dolde U, Wenkel S, Masakapalli SK, Datta S (2019a). The B- box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol 179, 1876-1892. |
| [77] | Yadav A, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2019b). BBX31 promotes hypocotyl growth, primary root elongation and UV-B tolerance in Arabidopsis. Plant Signal Behav 14, e1588672. |
| [78] |
Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2020). Light signaling and UV-B-mediated plant growth regulation. J Integr Plant Biol 62, 1270-1292.
DOI |
| [79] | Yadukrishnan P, Datta S (2021). Light and abscisic acid interplay in early seedling development. New Phytol 229, 763-769. |
| [80] | Yan HL, Pei XN, Zhang H, Li X, Zhang XX, Zhao MH, Chiang VL, Sederoff RR, Zhao XY (2021). MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci 22, 3103. |
| [81] | Yang GQ, Zhang CL, Dong HX, Liu XR, Guo HC, Tong BQ, Fang F, Zhao YY, Yu YJ, Liu Y, Lin L, Yin RH (2022). Activation and negative feedback regulation of SlHY5 transcription by the SlBBX20/21-SlHY5 transcription factor module in UV-B signaling. Plant Cell 34, 2038-2055. |
| [82] | Yang Y, Liang T, Zhang LB, Shao K, Gu XX, Shang RX, Shi N, Li X, Zhang P, Liu HT (2018). UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4, 98-107. |
| [83] | Yang Y, Liu HT (2020). Coordinated shoot and root responses to light signaling in Arabidopsis. Plant Commun 1, 100026. |
| [84] | Yang Y, Zhang LB, Chen P, Liang T, Li X, Liu HT (2020). UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39, e101928. |
| [85] | Yao JW, Ma Z, Ma YQ, Zhu Y, Lei MQ, Hao CY, Chen LY, Xu ZQ, Huang X (2021). Role of melatonin in UV-B signaling pathway and UV-B stress resistance in Arabidopsis thaliana. Plant Cell Environ 44, 114-129. |
| [86] |
Yin RH, Skvortsova MY, Loubéry S, Ulm R (2016). COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113, 4415-4422.
DOI PMID |
| [87] | Yin XJ, Zhang YB, Zhang L, Wang BH, Zhao YD, Irfan M, Chen LJ, Feng YL (2021). Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front Plant Sci 12, 761668. |
| [88] | Zeng YW, Schotte S, Trinh HK, Verstraeten I, Li J, Van De velde E, Vanneste S, Geelen D (2022). Genetic dissection of light-regulated adventitious root induction in Arabidopsis thaliana hypocotyls. Int J Mol Sci 23, 5301. |
| [89] | Zhang QW, Lin L, Fang F, Cui BM, Zhu C, Luo SK, Yin RH (2023). Dissecting the functions of COP1 in the UVR8 pathway with a COP1variant in Arabidopsis. Plant J 113, 478-492. |
| [90] |
Zhang XY, Huai JL, Shang FF, Xu G, Tang WJ, Jing YJ, Lin RC (2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174, 2487-2500.
DOI PMID |
| [91] | Zhang ZH, Xu CJ, Zhang SY, Shi C, Cheng H, Liu HT, Zhong BJ (2022). Origin and adaptive evolution of UV RESISTANCE LOCUS 8-mediated signaling during plant terrestrialization. Plant Physiol 188, 332-346. |
| [92] | Zhao JY, Bo KL, Pan YP, Li YH, Yu DL, Li C, Chang J, Wu S, Wang ZY, Zhang XL, Gu XF, Weng YQ (2023). Phytochrome-interacting factor PIF3 integrates phytochrome B and UV-B signaling pathways to regulate gibberellin- and auxin-dependent growth in cucumber hypocotyls. J Exp Bot 74, 4520-4539. |
| [93] | Zhou H, Zhu W, Wang XC, Bian YT, Jiang Y, Li J, Wang LX, Yin P, Deng XW, Xu DQ (2022a). A missense mutation in WRKY32 converts its function from a positive regulator to a repressor of photomorphogenesis. New Phytol 235, 111-125. |
| [94] | Zhou J, Meng JX, Zhang SY, Chi RF, Wang C, Wang DM, Li HH (2022b). The UV-B-induced transcription factor HY5 regulated anthocyanin biosynthesis in Zanthoxylum bungeanum. Int J Mol Sci 23, 2651. |
| [1] | 江亚楠, 徐雨青, 魏毅铤, 陈钧, 张蓉菀, 赵蓓蓓, 林宇翔, 饶玉春. 水稻抗病调控机制研究进展[J]. 植物学报, 2025, 60(5): 1-0. |
| [2] | 王笑, 徐昌文, 钱虹萍, 李思博, 林金星, 崔亚宁. 植物细胞壁参与免疫反应的机制及其原位非标记成像方法[J]. 植物学报, 2025, 60(5): 1-0. |
| [3] | 刘德水, 岳宁, 刘玉乐. 植物免疫机制新突破[J]. 植物学报, 2025, 60(5): 1-0. |
| [4] | 徐羽丰 周冕. 植物免疫的转录后调控[J]. 植物学报, 2025, 60(5): 1-0. |
| [5] | 谢涛, 章一帆, 刘云辉, 游慧玉, 夏季奔奔, 马蓉, 张春妮, 华学军. 植物线粒体铁硫簇合成系统及其调控的研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [6] | 魏梦璐, 李锦粤, 饶玉春. 基于“以赛促创”的植物科学领域创新型人才培养实践[J]. 植物学报, 2025, 60(4): 1-0. |
| [7] | 刘旭鹏, 王敏, 韩守安, 朱学慧, 王艳蒙, 潘明启, 张雯. 植物器官脱落调控因素及分子机理研究进展[J]. 植物学报, 2025, 60(3): 472-482. |
| [8] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [9] | 吕加一, 李乐攻, 侯聪聪. 基于FRET原理的生物传感器: 小分子荧光探针在植物中的研究进展[J]. 植物学报, 2025, 60(2): 283-293. |
| [10] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [11] | 顾红雅, 陈凡, 林荣呈, 漆小泉, 杨淑华, 陈之端, 陈学伟, 丁兆军, 萧浪涛, 左建儒, 姜里文, 白永飞, 种康, 王雷. 2024年中国植物科学重要研究进展[J]. 植物学报, 2025, 60(2): 151-171. |
| [12] | 邓言, 鲁丽敏, 张强, 陈之端, 胡海花. 维管植物质体DNA数据在物种和区域上的空缺研究(长英文摘要)[J]. 植物学报, 2025, 60(1): 1-16. |
| [13] | 陈秀秀, 唐玲, 胡文佳, 杨照麟, 邓馨, 王晓华. 植物细胞质膜有序性的活细胞定量分析[J]. 植物学报, 2025, 60(1): 90-100. |
| [14] | 宣晶, 付其迪, 谢淦, 薛凯, 雒海瑞, 魏泽, 赵明月, 智亮, 万华伟, 高吉喜, 李敏. 北方草地植物物种智能识别模型构建及应用[J]. 植物学报, 2025, 60(1): 74-80. |
| [15] | 王亚萍, 包文泉, 白玉娥. 单细胞转录组学在植物生长发育及胁迫响应中的应用进展[J]. 植物学报, 2025, 60(1): 101-113. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||