

植物学报 ›› 2025, Vol. 60 ›› Issue (1): 1-16.DOI: 10.11983/CBB24034 cstr: 32102.14.CBB24034
• 研究论文 • 下一篇
邓言1,2, 鲁丽敏2,3, 张强4,*( ), 陈之端2,3, 胡海花2,3,*(
), 陈之端2,3, 胡海花2,3,*( )
)
收稿日期:2024-03-06
接受日期:2024-05-27
出版日期:2025-01-10
发布日期:2024-05-27
通讯作者:
胡海花, 中国科学院植物研究所副研究员。长期从事维管植物生命之树重建及生物多样性格局与保护研究。已发表研究论文20余篇, 其中以第一作者(含共同第一作者)身份在National Science Review、Fundamental Research、Journal of Systematics and Evolution等期刊发表研究论文4篇。所在的植物大数据与生物多样性保护研究团队利用多学科手段, 在较高分类阶元上探讨植物的系统发育关系和演化, 并将形态学、古植物学和分子系统学的研究结果相结合, 研究植物类群的起源、分化和现代地理分布格局及其成因。近年来, 以生命之树为依托, 结合海量物种分布数据, 从时间和空间维度探究植物区系的演化历史、多样性格局及其成因及生物多样性保护策略。E-mail: 基金资助:
Yan Deng1,2, Limin Lu2,3, Qiang Zhang4,*( ), Zhiduan Chen2,3, Haihua Hu2,3,*(
), Zhiduan Chen2,3, Haihua Hu2,3,*( )
)
Received:2024-03-06
Accepted:2024-05-27
Online:2025-01-10
Published:2024-05-27
Contact:
* E-mail: 摘要: 在植物大数据时代, 测序数据成为众多生物学研究的重要基础, 了解测序数据的现状有利于更好地利用这些数据。质体DNA数据因易获取、单亲遗传及变异速率适中而被广泛应用。基于GenBank公共数据库全面评估和分析了全世界维管植物质体DNA数据取样情况, 结果表明, 仅有33.75%的维管植物种类已测序。已测序物种在不同类群间取样不均衡, 缺失率大致与类群多样性呈显著正相关, 其中缺失最严重的目和科分别是盔被花目(Paracryphiales)、胡椒目(Piperales)和五桠果目(Dilleniales), 以及霉草科(Triuridaceae)、五膜草科(Pentaphragmataceae)和黄眼草科(Xyridaceae)。在地理空间上, 维管植物数据缺失程度从赤道向两极递减, 且生物多样性高的地区缺失更严重, 包括多个生物多样性热点地区。此外, 各地区特有种的数据普遍缺失严重。基于上述结果, 建议针对分子数据缺失程度较高的类群和生物多样性高的地区进行重点采集和测序, 尤其注重对特有种补充取样, 以增加这些类群遗传数据的代表性。
邓言, 鲁丽敏, 张强, 陈之端, 胡海花. 维管植物质体DNA数据在物种和区域上的空缺研究(长英文摘要). 植物学报, 2025, 60(1): 1-16.
Yan Deng, Limin Lu, Qiang Zhang, Zhiduan Chen, Haihua Hu. A Comprehensive Evaluation of the Plastid DNA Data Gaps of Vascular Plants in Species and Geographic Area. Chinese Bulletin of Botany, 2025, 60(1): 1-16.
| Group | Number of sampled species | Sampling ratio (%) |
|---|---|---|
| Tracheophytes | 139005 | 33.75 |
| Angiosperms | 131220 | 33.16 |
| Gymnosperms | 1154 | 75.92 |
| Pteridophytes | 6631 | 45.35 |
表1 维管植物、被子植物、裸子植物和广义蕨类植物的取样物种数和取样率
Table 1 Number of sampled species and sampling ratios in tracheophytes, angiosperms, gymnosperms, and pteridophytes
| Group | Number of sampled species | Sampling ratio (%) |
|---|---|---|
| Tracheophytes | 139005 | 33.75 |
| Angiosperms | 131220 | 33.16 |
| Gymnosperms | 1154 | 75.92 |
| Pteridophytes | 6631 | 45.35 |
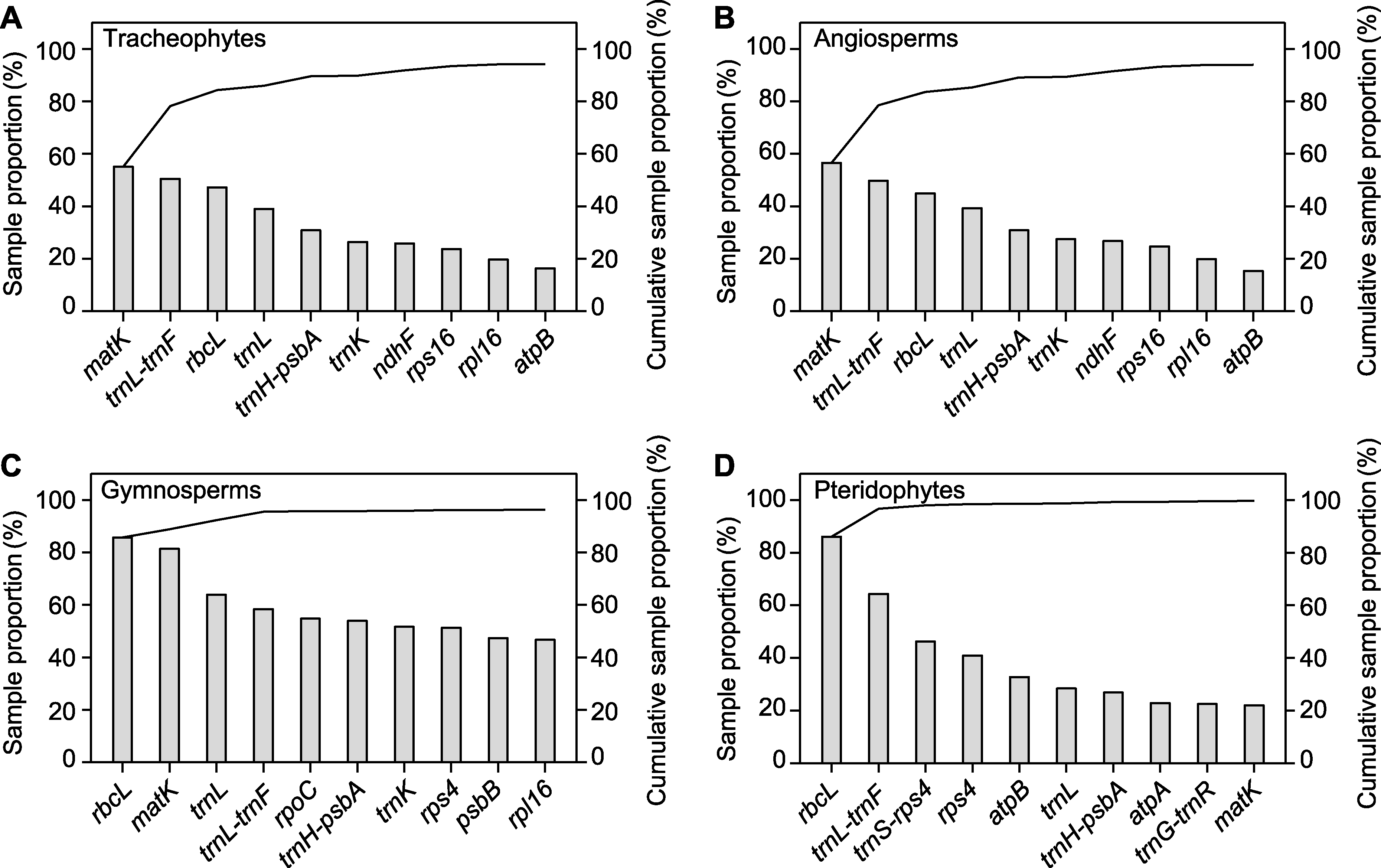
图1 GenBank中维管植物(A)、被子植物(B)、裸子植物(C)和广义蕨类植物(D)取样率排名前10的质体DNA分子标记的取样率及累积取样率 柱状图表示每个分子标记的取样率, 黑色实线表示累积取样率。
Figure 1 Sample proportion and cumulative sample proportion of the top ten plastid DNA molecular markers in GenBank for tracheophytes (A), angiosperms (B), gymnosperms (C), and pteridophytes (D) Histograms represent the sample proportion of each molecular marker, and black lines represent the cumulative sample proportion.
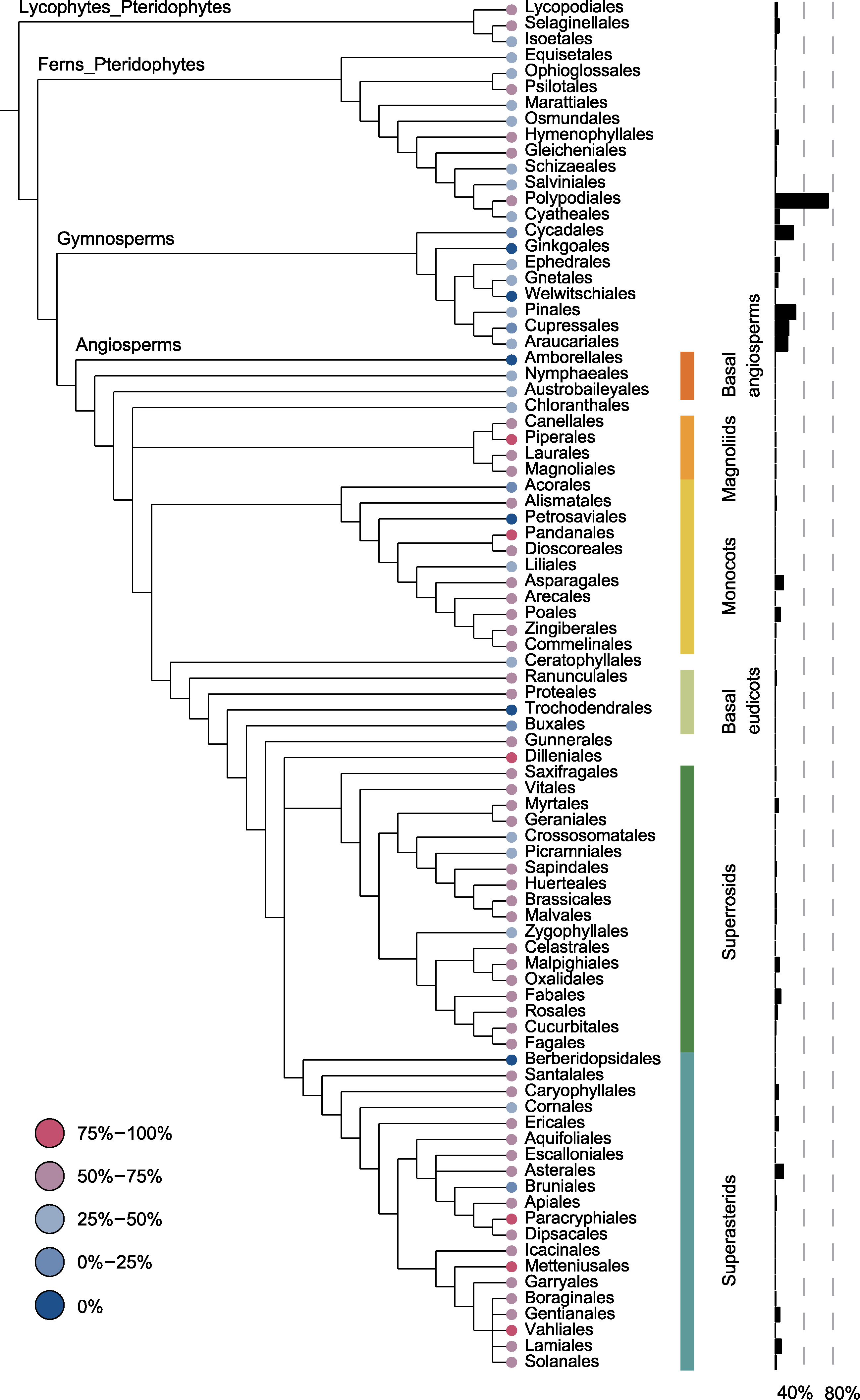
图2 维管植物目水平质体DNA数据缺失情况和每个目的多样性占比 系统发生树末端节点的颜色表示缺失情况, 右侧的黑色柱状图表示每个目分别在被子植物、裸子植物和广义蕨类植物中的多样性占比。维管植物系统树改自APG IV (The Angiosperm Phylogeny Group et al., 2016)、Christenhusz (Christenhusz et al., 2011)和PPG I (PPG I, 2016)。被子植物主要分支用不同颜色条带表示。
Figure 2 Missing data in plastid DNA and the proportions of species diversity at the ordinal level of tracheophytes Colored circles at the terminal of phylogenetic tree represent the proportion of missing data in plastid DNA and black histograms on the right represent the proportion of species diversity of each order in angiosperms, gymnosperms, and pteridophytes, respectively. The tracheophyte phylogenetic tree was modified from APG IV (The Angiosperm Phylogeny Group et al., 2016), Christenhusz (Christenhusz et al., 2011), and PPG I (PPG I, 2016). The major clades of angiosperms are indicated with bars of different colors.
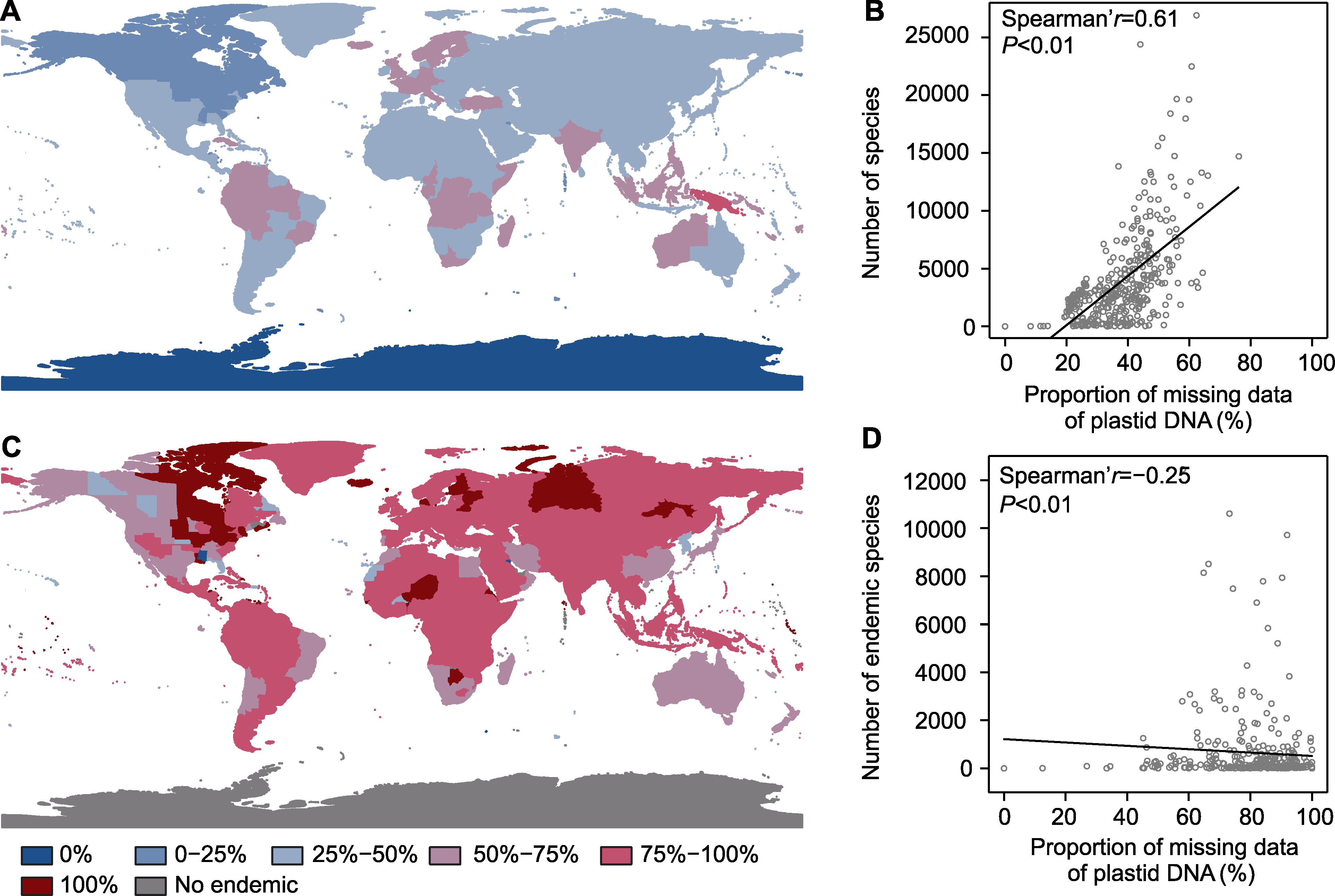
图3 全球维管植物(A)和特有维管植物(C)的质体DNA数据缺失情况的空间格局以及地区物种多样性(B)和特有种多样性(D)分别与各自的质体DNA数据缺失率的Spearman相关性
Figure 3 Spatial patterns of missing data in plastid DNA of global tracheophytes (A) and endemic tracheophytes (C), and Spearman correlations between regional species diversity and missing data in plastid DNA for tracheophytes (B) and endemic tracheophytes (D), respectively.
| [1] |
Burbano HA, Gutaker RM (2023). Ancient DNA genomics and the renaissance of herbaria. Science 382, 59-63.
DOI PMID |
| [2] | Cámara-Leret R, Frodin DG, Adema F, Anderson C, Appelhans MS, Argent G, Arias Guerrero S, Ashton P, Baker WJ, Barfod AS, Barrington D, Borosova R, Bramley GLC, Briggs M, Buerki S, Cahen D, Callmander MW, Cheek M, Chen CW, Conn BJ, Coode MJE, Darbyshire I, Dawson S, Dransfield J, Drinkell C, Duyfjes B, Ebihara A, Ezedin Z, Fu LF, Gideon O, Girmansyah D, Govaerts R, Fortune-Hopkins H, Hassemer G, Hay A, Heatubun CD, Hind DJN, Hoch P, Homot P, Hovenkamp P, Hughes M, Jebb M, Jennings L, Jimbo T, Kessler M, Kiew R, Knapp S, Lamei P, Lehnert M, Lewis GP, Linder HP, Lindsay S, Low YW, Lucas E, Mancera JP, Monro AK, Moore A, Middleton DJ, Nagamasu H, Newman MF, Nic Lughadha E, Melo PHA, Ohlsen DJ, Pannell CM, Parris B, Pearce L, Penneys DS, Perrie LR, Petoe P, Poulsen AD, Prance GT, Quakenbush JP, Raes N, Rodda M, Rogers ZS, Schuiteman A, Schwartsburd P, Scotland RW, Simmons MP, Simpson DA, Stevens P, Sundue M, Testo W, Trias-Blasi A, Turner I, Utteridge T, Walsingham L, Webber BL, Wei R, Weiblen GD, Weigend M, Weston P, de Wilde W, Wilkie P, Wilmot-Dear CM, Wilson HP, Wood JRI, Zhang LB, van Welzen PC (2020). New Guinea has the world’s richest island flora. Nature 584, 579-583. |
| [3] | Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu YL, Kron KA, Rettig JH, Conti E, Palmer JD, Manhart JR, Sytsma KJ, Michaels HJ, Kress WJ, Karol KG, Clark WD, Hedren M, Gaut BS, Jansen RK, Kim KJ, Wimpee CF, Smith JF, Furnier GR, Strauss SH, Xiang QY, Plunkett GM, Soltis PS, Swensen SM, Williams SE, Gadek PA, Quinn CJ, Eguiarte LE, Golenberg E, Learn GH, Graham SW, Barrett SCH, Dayanandan S, Albert VA (1993). Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard 80, 528-548, 550-580. |
| [4] |
Chen QH, Chen L, Teixeira da Silva JA, Yu XN (2023). The plastome reveals new insights into the evolutionary and domestication history of peonies in East Asia. BMC Plant Biol 23, 243.
DOI PMID |
| [5] | Chen ZD, Yang T, Lin L, Lu LM, Li HL, Sun M, Liu B, Chen M, Niu YT, Ye JF, Cao ZY, Liu HM, Wang XM, Wang W, Zhang JB, Meng Z, Cao W, Li JH, Wu SD, Zhao HL, Liu ZJ, Du ZY, Wang QF, Guo J, Tan XX, Su JX, Zhang LJ, Yang LL, Liao YY, Li MH, Zhang GQ, Chung SW, Zhang J, Xiang KL, Li RQ, Soltis DE, Soltis PS, Zhou SL, Ran JH, Wang XQ, Jin XH, Chen YS, Gao TG, Li JH, Zhang SZ, Lu AM, China Phylogeny Consortium (2016). Tree of life for the genera of Chinese vascular plants. J Syst Evol 54, 277-306. |
| [6] | Christenhusz MJM, Reveal JL, Farjon A, Gardner MF, Mill RR, Chase MW (2011). A new classification and linear sequence of extant gymnosperms. Phytotaxa 19, 55-70. |
| [7] | Christin PA, Spriggs E, Osborne CP, Strömberg CAE, Salamin N, Edwards EJ (2014). Molecular dating, evolutionary rates, and the age of the grasses. Syst Biol 63, 153-165. |
| [8] | Coe K, Bostan H, Rolling W, Turner-Hissong S, Macko- Podgórni A, Senalik D, Liu S, Seth R, Curaba J, Mengist MF, Grzebelus D, Van Deynze A, Dawson J, Ellison S, Simon P, Iorizzo M (2023). Population genomics identifies genetic signatures of carrot domestication and improvement and uncovers the origin of high- carotenoid orange carrots. Nat Plants 9, 1643-1658. |
| [9] | Coelho N, Gonçalves S, Romano A (2020). Endemic plant species conservation: biotechnological approaches. Plants (Basel) 9, 345. |
| [10] |
Cornwell WK, Pearse WD, Dalrymple RL, Zanne AE (2019). What we (don’t) know about global plant diversity. Ecography 42, 1819-1831.
DOI |
| [11] |
Folk RA, Sun M, Soltis PS, Smith SA, Soltis DE, Guralnick RP (2018). Challenges of comprehensive taxon sampling in comparative biology: wrestling with rosids. Am J Bot 105, 433-445.
DOI PMID |
| [12] | Forest F, Grenyer R, Rouget M, Davies TJ, Cowling RM, Faith DP, Balmford A, Manning JC, Procheş Ş, van der Bank M, Reeves G, Hedderson TAJ, Savolainen V (2007). Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757-760. |
| [13] |
Goremykin VV, Hirsch-Ernst KI, Wölfl S, Hellwig FH (2003). Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol 20, 1499-1505.
PMID |
| [14] |
Govaerts R, Nic Lughadha E, Black N, Turner R, Paton A (2021). The world checklist of vascular plants, a continuously updated resource for exploring global plant diversity. Sci Data 8, 215.
DOI PMID |
| [15] |
Gutaker RM, Burbano HA (2017). Reinforcing plant evolutionary genomics using ancient DNA. Curr Opin Plant Biol 36, 38-45.
DOI PMID |
| [16] | He JK, Gao ZF, Su YY, Lin SL, Jiang HS (2018). Geographical and temporal origins of terrestrial vertebrates endemic to Taiwan. J Biogeogr 45, 2458-2470. |
| [17] | Hinchliff CE, Smith SA (2014). Some limitations of public sequence data for phylogenetic inference (in plants). PLoS One 9, e98986. |
| [18] |
Hu HH, Liu B, Liang YS, Ye JF, Saqib S, Meng Z, Lu LM, Chen ZD (2020). An updated Chinese vascular plant tree of life: phylogenetic diversity hotspots revisited. J Syst Evol 58, 663-672.
DOI |
| [19] | Hu HH, Ye JF, Liu B, Mao LF, Smith SA, Barrett RL, Soltis PS, Soltis DE, Chen ZD, Lu LM (2022). Temporal and spatial comparisons of angiosperm diversity between eastern Asia and North America. Natl Sci Rev 9, nwab 199. |
| [20] |
Hu Y, Wang X, Zhang XX, Zhou W, Chen XY, Hu XS (2019). Advancing phylogeography with chloroplast DNA markers. Biodiv Sci 27, 219-234. (in Chinese)
DOI |
|
胡颖, 王茜, 张新新, 周玮, 陈晓阳, 胡新生 (2019). 叶绿体DNA标记在谱系地理学中的应用研究进展. 生物多样性 27, 219-234.
DOI |
|
| [21] | Huang WD, Zhao XY, Zhao X, Li YQ, Zuo XA, Lian J, Luo YQ (2013). Genetic diversity in Artemisia halodendron (Asteraceae) based on chloroplast DNA psbA-trnH region from different hydrothermal conditions in Horqin sandy land, northern China. Plant Syst Evol 299, 107-113. |
| [22] |
Iorizzo M, Ellison S, Senalik D, Zeng P, Satapoomin P, Huang JY, Bowman M, Iovene M, Sanseverino W, Cavagnaro P, Yildiz M, Macko-Podgórni A, Moranska E, Grzebelus E, Grzebelus D, Ashrafi H, Zheng ZJ, Cheng SF, Spooner D, Van Deynze A, Simon P (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet 48, 657-666.
DOI PMID |
| [23] | Jafari F, Osaloo SK, Mozffarian V (2015). Molecular phylogeny of the tribe Astereae (Asteraceae) in SW Asia based on nrDNA ITS and cpDNA psbA-trnH sequences. Willdenowia 45, 77-92. |
| [24] | Janssens SB, Couvreur TLP, Mertens A, Dauby G, Dagallier LPMJ, Vanden Abeele S, Vandelook F, Mascarello M, Beeckman H, Sosef M, Droissart V, van der Bank M, Maurin O, Hawthorne W, Marshall C, Réjou- Méchain M, Beina D, Baya F, Merckx V, Verstraete B, Hardy O (2020). A large-scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. Biodivers Data J 8, e39677. |
| [25] | Jiang ZH, Zhang MQ, Kong LY, Bao YH, Ren WC, Li HY, Liu XB, Wang Z, Ma W (2023). Identification of Apiaceae using ITS, ITS2 and psbA-trnH barcodes. Mol Biol Rep 50, 245-253. |
| [26] |
Kistler L, Bieker VC, Martin MD, Pedersen MW, Ramos Madrigal J, Wales N (2020). Ancient plant genomics in archaeology, herbaria, and the environment. Annu Rev Plant Biol 71, 605-629.
DOI PMID |
| [27] | Krause CM, Cobb NS, Pennington DD (2015). Range shifts under future scenarios of climate change: dispersal ability matters for Colorado plateau endemic plants. Nat Areas J 35, 428-438. |
| [28] | Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L (2011). Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome 54, 663-673. |
| [29] | Lang PLM, Weiß CL, Kersten S, Latorre SM, Nagel S, Nickel B, Meyer M, Burbano HA (2020). Hybridization ddRAD-sequencing for population genomics of nonmodel plants using highly degraded historical specimen DNA. Mol Ecol Resour 20, 1228-1247. |
| [30] | Li FD, Tong W, Xia EH, Wei CL (2019). Optimized sequencing depth and de novo assembler for deeply reconstructing the transcriptome of the tea plant, an economically important plant species. BMC Bioinformatics 20, 553. |
| [31] | Li HT, Luo Y, Gan L, Ma PF, Gao LM, Yang JB, Cai J, Gitzendanner MA, Fritsch PW, Zhang T, Jin JJ, Zeng CX, Wang H, Yu WB, Zhang R, van der Bank M, Olmstead RG, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Yi TS, Li DZ (2021). Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol 19, 232. |
| [32] | Li L, Wang WY, Zhang GQ, Wu KL, Fang L, Li MZ, Liu ZJ, Zeng SJ (2023). Comparative analyses and phylogenetic relationships of thirteen Pholidota species (Orchidaceae) inferred from complete chloroplast genomes. BMC Plant Biol 23, 269. |
| [33] | Lian L, Peng HW, Ortiz RDC, Jabbour F, Gao TG, Erst AS, Chen ZD, Wang W (2023). Phylogeny and biogeography of Tiliacoreae (Menispermaceae), a tribe restricted to tropical rainforests. Ann Bot 131, 685-695. |
| [34] |
Linder HP, Hardy CR, Rutschmann F (2005). Taxon sampling effects in molecular clock dating: an example from the African Restionaceae. Mol Phylogenet Evol 35, 569-582.
PMID |
| [35] | Liu YJ, Liu Y, Huang YJ, Long CL (2011). Progress and application of DNA barcoding technique in plants. J Plant Resour Environ 20, 74-82, 93. (in Chinese) |
| 刘宇婧, 刘越, 黄耀江, 龙春林 (2011). 植物DNA条形码技术的发展及应用. 植物资源与环境学报 20, 74-82, 93. | |
| [36] | Liu YM, Zhang LH, Liu Z, Luo K, Chen SL, Chen KL (2012). Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid psbA- trnH genetic marker. Pharmacogn Mag 8, 29-36. |
| [37] |
Liu YP, Xu XT, Dimitrov D, Pellissier L, Borregaard MK, Shrestha N, Su XY, Luo A, Zimmermann NE, Rahbek C, Wang ZH (2023). An updated floristic map of the world. Nat Commun 14, 2990.
DOI PMID |
| [38] | Lu LM, Mao LF, Yang T, Ye JF, Liu B, Li HL, Sun M, Miller JT, Mathews S, Hu HH, Niu YT, Peng DX, Chen YH, Smith SA, Chen M, Xiang KL, Le CT, Dang VC, Lu AM, Soltis PS, Soltis DE, Li JH, Chen ZD (2018). Evolutionary history of the angiosperm flora of China. Nature 554, 234-238. |
| [39] | Lv YN, Yang CY, Shi LC, Zhang ZL, Xu AS, Zhang LX, Li XL, Li HT (2020). Identification of medicinal plants within the Apocynaceae family using ITS2 and psbA-trnH barcodes. Chin J Nat Med 18, 594-605. |
| [40] | Mo ZQ, Fu CN, Zhu MS, Milne RI, Yang JB, Cai J, Qin HT, Zheng W, Hollingsworth PM, Li DZ, Gao LM (2022). Resolution, conflict and rate shifts: insights from a densely sampled plastome phylogeny for Rhododendron (Ericaceae). Ann Bot 130, 687-701. |
| [41] | Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853-858. |
| [42] |
Nock CJ, Waters DLE, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ (2011). Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J 9, 328-333.
DOI PMID |
| [43] |
Novikova PY, Hohmann N, Nizhynska V, Tsuchimatsu T, Ali J, Muir G, Guggisberg A, Paape T, Schmid K, Fedorenko OM, Holm S, Säll T, Schlötterer C, Marhold K, Widmer A, Sese J, Shimizu KK, Weigel D, Krämer U, Koch MA, Nordborg M (2016). Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet 48, 1077-1082.
DOI PMID |
| [44] | Ohi T, Kajita T, Murata J (2003). Distinct geographic structure as evidenced by chloroplast DNA haplotypes and ploidy level in Japanese Aucuba (Aucubaceae). Am J Bot 90, 1645-1652. |
| [45] |
Ortelt J, Link G (2014). Plastid gene transcription: promoters and RNA polymerases. Methods Mol Biol 1132, 47-72.
DOI PMID |
| [46] |
Park DS, Worthington S, Xi ZX (2018). Taxon sampling effects on the quantification and comparison of community phylogenetic diversity. Mol Ecol 27, 1296-1308.
DOI PMID |
| [47] | Peng HW, Wang W (2023). Phylogenetic tree reconstruction based on molecular data. Chin Bull Bot 58, 261-273. (in Chinese) |
|
彭焕文, 王伟 (2023). 基于分子数据的系统发生树构建. 植物学报 58, 261-273.
DOI |
|
| [48] |
PPG I (2016). A community-derived classification for extant lycophytes and ferns. J Syst Evol 54, 563-603.
DOI |
| [49] | Raman G, Park S (2015). Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS One 10, e0141329. |
| [50] | Ran JH, Shen TT, Wang MM, Wang XQ (2018). Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc Biol Sci 285, 20181012. |
| [51] | Rudbeck AV, Sun M, Tietje M, Gallagher RV, Govaerts R, Smith SA, Svenning JC, Eiserhardt WL (2022). The Darwinian shortfall in plants: phylogenetic knowledge is driven by range size. Ecography 2022, e06142. |
| [52] | Schulte JA (2013). Undersampling taxa will underestimate molecular divergence dates: an example from the South American Lizard clade Liolaemini. Int J Evol Biol 2013, 628467. |
| [53] |
Smith SA, Brown JW (2018). Constructing a broadly inclusive seed plant phylogeny. Am J Bot 105, 302-314.
DOI PMID |
| [54] | Soltis DE, Soltis PS (2004). Amborella not a “basal angiosperm”? Not so fast. Am J Bot 91, 997-1001. |
| [55] | Staats M, Cuenca A, Richardson JE, Ginkel RVV, Petersen G, Seberg O, Bakker FT (2011). DNA damage in plant herbarium tissue. PLoS One 6, e28448. |
| [56] | The Angiosperm Phylogeny Group, Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181, 1-20. |
| [57] | Tyler T (2017). The last step towards a full revision of Hieracium sect. Vulgata in Sweden. Nord J Bot 35, 305-321. |
| [58] | Vischi N, Natale E, Villamil C (2004). Six endemic plant species from central Argentina: an evaluation of their conservation status. Biodivers Conserv 13, 997-1008. |
| [59] | Wang J, Kan SL, Liao XZ, Zhou JW, Tembrock LR, Daniell H, Jin SX, Wu ZQ (2024). Plant organellar genomes: much done, much more to do. Trends Plant Sci 29, 754-769. |
| [60] | Wang LL, Yang Z, Yang Y (2023). Plant ultra-barcoding using herbariomics. Chin Bull Bot 58, 831-842. (in Chinese) |
|
王露露, 杨智, 杨永 (2023). 利用标本组学推进植物超级DNA条形码研究. 植物学报 58, 831-842.
DOI |
|
| [61] |
Wicke S, Schneeweiss GM, de Pamphilis CW, Müller KF, Quandt D (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76, 273-297.
DOI PMID |
| [62] | Yao X, Song Y, Yang JB, Tan YH, Corlett RT (2021). Phylogeny and biogeography of the hollies (Ilex L., Aquifoliaceae). J Syst Evol 59, 73-82. |
| [63] | Ye JW, Yang ZZ, Tian B (2023). Tempo-spatial evolution of seed plant endemism in Taiwan island. J Biogeogr 50, 1981-1991. |
| [64] | Yu QB, Huang C, Yang ZN (2014). Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci 5, 316. |
| [65] | Yurina NP, Sharapova LS, Odintsova MS (2017). Structure of plastid genomes of photosynthetic eukaryotes. Biochemistry (Mosc) 82, 678-691. |
| [66] | Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM (2014). Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89-92. |
| [67] |
Zhang X, Sun YX, Landis JB, Lv ZY, Shen J, Zhang HJ, Lin N, Li LJ, Sun J, Deng T, Sun H, Wang HC (2020). Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol 20, 340.
DOI PMID |
| [68] | Zhang YJ, Li DZ (2011). Advances in phylogenomics based on complete chloroplast genomes. Plant Diversity Resour 33, 365-375. (in Chinese) |
| 张韵洁, 李德铢 (2011). 叶绿体系统发育基因组学的研究进展. 植物分类与资源学报 33, 365-375. |
| [1] | 郑博瀚, 陈鑫瑶, 倪健. 中国维管植物生长型和生活型数据集[J]. 生物多样性, 2024, 32(7): 23468-. |
| [2] | 万霞, 张丽兵. 世界维管植物新分类群2023年度报告[J]. 生物多样性, 2024, 32(11): 24322-. |
| [3] | 韩赟, 迟晓峰, 余静雅, 丁旭洁, 陈世龙, 张发起. 青海野生维管植物名录[J]. 生物多样性, 2023, 31(9): 23280-. |
| [4] | 陈又生, 宋柱秋, 卫然, 罗艳, 陈文俐, 杨福生, 高连明, 徐源, 张卓欣, 付鹏程, 向春雷, 王焕冲, 郝加琛, 孟世勇, 吴磊, 李波, 于胜祥, 张树仁, 何理, 郭信强, 王文广, 童毅华, 高乞, 费文群, 曾佑派, 白琳, 金梓超, 钟星杰, 张步云, 杜思怡. 西藏维管植物多样性编目和分布数据集[J]. 生物多样性, 2023, 31(9): 23188-. |
| [5] | 陈慧妹, 李文军, 邱娟, 马占仓, 李波, 杨宗宗, 闻志彬, 孟岩, 曹秋梅, 邱东, 刘丹辉, 金光照. 新疆野生维管植物名录[J]. 生物多样性, 2023, 31(9): 23124-. |
| [6] | 李勇, 李三青, 王欢. 天津野生维管植物编目及分布数据集[J]. 生物多样性, 2023, 31(9): 23128-. |
| [7] | 杜诚, 汪远, 闫小玲, 严靖, 李惠茹, 张庆费, 胡永红. 上海市植物物种多样性组成和历史变化暨上海维管植物名录更新(2022版)[J]. 生物多样性, 2023, 31(6): 23093-. |
| [8] | 安昌, 庄怡雪, 郑平, 林彦翔, 杨成梓, 秦源. 福建省维管植物名录[J]. 生物多样性, 2023, 31(6): 22537-. |
| [9] | 韦毅刚, 温放, 辛子兵, 符龙飞. 广西野生维管植物名录[J]. 生物多样性, 2023, 31(6): 23078-. |
| [10] | 梁彩群, 陈玉凯, 杨小波, 张凯, 李东海, 江悦馨, 李婧涵, 王重阳, 张顺卫, 朱子丞. 海南省野生维管植物编目和分布数据集[J]. 生物多样性, 2023, 31(6): 23067-. |
| [11] | 万霞, 张丽兵. 世界维管植物新分类群2022年度报告[J]. 生物多样性, 2023, 31(10): 23162-. |
| [12] | 万霞, 张丽兵. 世界维管植物新分类群2021年年度报告[J]. 生物多样性, 2022, 30(8): 22116-. |
| [13] | 王婷, 舒江平, 顾钰峰, 李艳清, 杨拓, 徐洲锋, 向建英, 张宪春, 严岳鸿. 中国石松类和蕨类植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22381-. |
| [14] | 寄玲, 谢宜飞, 李中阳, 许廷晨, 杨波, 李波. 江西省野生维管植物名录[J]. 生物多样性, 2022, 30(6): 22057-. |
| [15] | 王洪峰, 董雪云, 穆立蔷. 黑龙江省野生维管植物名录[J]. 生物多样性, 2022, 30(6): 22184-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||