

植物学报 ›› 2025, Vol. 60 ›› Issue (2): 283-293.DOI: 10.11983/CBB24056 cstr: 32102.14.CBB24056
收稿日期:2024-04-15
接受日期:2024-10-14
出版日期:2025-03-10
发布日期:2024-10-16
通讯作者:
侯聪聪
基金资助:
Jiayi Lü, Legong Li, Congcong Hou*( )
)
Received:2024-04-15
Accepted:2024-10-14
Online:2025-03-10
Published:2024-10-16
Contact:
Congcong Hou
摘要: 生物小分子是指生物体内分子量较小的单体物质, 植物小分子种类繁多, 包括离子、植物激素和代谢物等。了解植物体内这些小分子的动态变化, 有助于解析相关的生理功能和调控网络, 并为植物细胞学的精确观察创造新的机遇。基于Förster共振能量转移(Förster resonance energy transfer, FRET)原理设计的基因编码荧光生物传感器/探针, 为活体内观察这些小分子的动态变化提供了强有力的工具。通过FRET传感器/探针, 能够可视化细胞内特定小分子化合物的浓度, 并实时获取高分辨率图像。这一技术因其独特的优势而被广泛应用于植物生理学、发育生物学和环境科学等研究领域。该文总结了近年来植物学研究中使用的FRET传感器/探针, 概述了它们的主要设计思路, 并阐述了其在检测离子、植物激素及代谢物方面的应用与研究进展, 旨在为植物中生物小分子的功能研究提供实用的技术手段和可能的研究方向。
吕加一, 李乐攻, 侯聪聪. 基于FRET原理的生物传感器: 小分子荧光探针在植物中的研究进展. 植物学报, 2025, 60(2): 283-293.
Jiayi Lü, Legong Li, Congcong Hou. FRET-based Biosensors: Application of Small Molecule Fluorescence Probes in Plants. Chinese Bulletin of Botany, 2025, 60(2): 283-293.
| 生物传感器/探针 | 目标小分子 | 感知元件 | FRET供体 | FRET受体 | 参考文献 |
|---|---|---|---|---|---|
| YC | Ca2+ | CaM (calmodulin)-M13 | eCFP (enhanced cyan fluorescent protein) | eYFP (enhanced yellow fluorescent protein) | Miyawaki et al., |
| CALWY | Zn2+ | ATOX1和WD4 | eCFP | eYFP | van Dongen et al., |
| eCALWY | Zn2+ | ATOX1和WD4 | Cerulean | Citrine | Vinkenborg et al., |
| NiTrac1 | NO3- | NRT1.1 (nitrate transporter 1.1) | mCerulean | Aphrodite | Ho and Frommer, |
| NitraMeter3.0 | NO3- | NasR蛋白中的硝酸盐和亚硝酸盐结合结构域 | edeCFP | edAFP | Chen et al., |
| FLIPPi | Pi | PiBP (phosphate-binding protein) | eCFP | eYFP/cpVenus | Gu et al., |
| ABAleon | 脱落酸 | 全长PYR1 (pyrabactin resistance 1)和截短的ABI1 (ABA insensitive 1) | mTurquoise | cpVenus73 | Waadt et al., |
| ABACUS | 脱落酸 | 全长PYL1 (PYR-like protein 1)和ABI1催化结构域 | edCerulean | edCitrine | Jones et al., |
| GPS1 | 赤霉素 | 截短的GAI (gibberellic acid insensitive)和全长GID1C (GA insensitive dwarf 1c) | edCerulean | edAFP | Rizza et al., |
| AuxSen | 生长素 | TrpR (tryptophan repressor) | Aquamarine | mNeonGreen | Herud-Sikimić et al., |
| FLIPglus | 葡萄糖 | GGBP (glucose/galactose-binding protein) | eCFP | eYFP | Fehr et al., |
| Ateam | ATP | FoF1-ATP合酶的ε亚基 | mseCFP | cp173-mVenus | Imamura et al., |
| - | 谷氨酰胺 | QBP (glutamine binding protein) | CFP | YFP | Yang et al., |
表1 植物中基于Förster共振能量转移(FRET)原理的小分子生物传感器/探针
Table1 The Förster resonance energy transfer (FRET)-based biosensors/probes for small molecules in plants
| 生物传感器/探针 | 目标小分子 | 感知元件 | FRET供体 | FRET受体 | 参考文献 |
|---|---|---|---|---|---|
| YC | Ca2+ | CaM (calmodulin)-M13 | eCFP (enhanced cyan fluorescent protein) | eYFP (enhanced yellow fluorescent protein) | Miyawaki et al., |
| CALWY | Zn2+ | ATOX1和WD4 | eCFP | eYFP | van Dongen et al., |
| eCALWY | Zn2+ | ATOX1和WD4 | Cerulean | Citrine | Vinkenborg et al., |
| NiTrac1 | NO3- | NRT1.1 (nitrate transporter 1.1) | mCerulean | Aphrodite | Ho and Frommer, |
| NitraMeter3.0 | NO3- | NasR蛋白中的硝酸盐和亚硝酸盐结合结构域 | edeCFP | edAFP | Chen et al., |
| FLIPPi | Pi | PiBP (phosphate-binding protein) | eCFP | eYFP/cpVenus | Gu et al., |
| ABAleon | 脱落酸 | 全长PYR1 (pyrabactin resistance 1)和截短的ABI1 (ABA insensitive 1) | mTurquoise | cpVenus73 | Waadt et al., |
| ABACUS | 脱落酸 | 全长PYL1 (PYR-like protein 1)和ABI1催化结构域 | edCerulean | edCitrine | Jones et al., |
| GPS1 | 赤霉素 | 截短的GAI (gibberellic acid insensitive)和全长GID1C (GA insensitive dwarf 1c) | edCerulean | edAFP | Rizza et al., |
| AuxSen | 生长素 | TrpR (tryptophan repressor) | Aquamarine | mNeonGreen | Herud-Sikimić et al., |
| FLIPglus | 葡萄糖 | GGBP (glucose/galactose-binding protein) | eCFP | eYFP | Fehr et al., |
| Ateam | ATP | FoF1-ATP合酶的ε亚基 | mseCFP | cp173-mVenus | Imamura et al., |
| - | 谷氨酰胺 | QBP (glutamine binding protein) | CFP | YFP | Yang et al., |
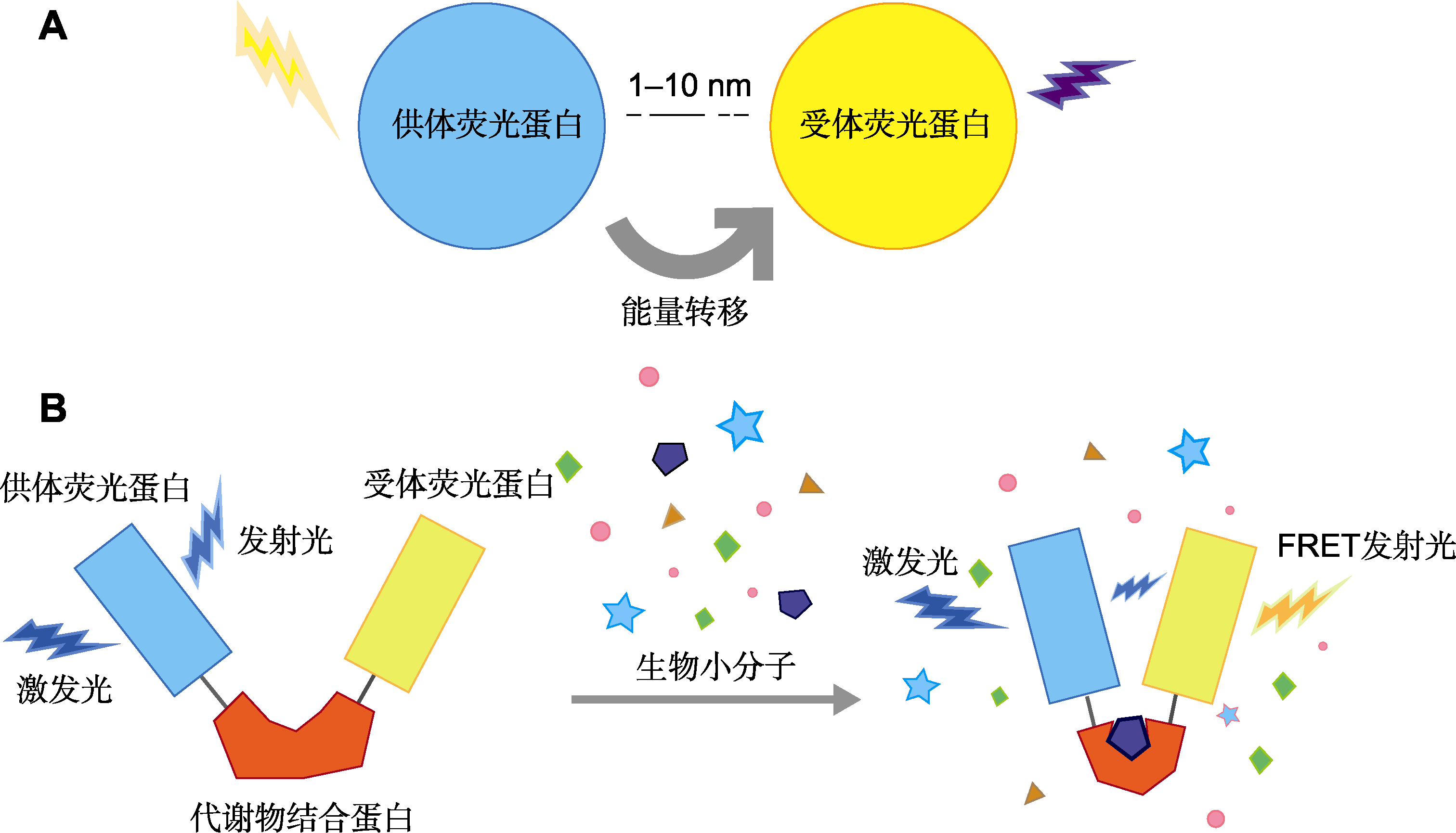
图1 基于Förster共振能量转移(FRET)原理的生物传感器/探针 (A) FRET原理; (B) 基于FRET原理设计的生物传感器/探针结构
Figure1 Förster resonance energy transfer (FRET)-based biosensors/probes (A) Principle of FRET; (B) Structure of FRET-based biosensors/probes
| [1] | Balcerowicz M, Shetty KN, Jones AM (2021). Fluorescent biosensors illuminating plant hormone research. Plant Phy- siol 187, 590-602. |
| [2] | Behera S, Wang NL, Zhang CX, Schmitz-Thom I, Strohkamp S, Schültke S, Hashimoto K, Xiong LZ, Kudla J (2015). Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytol 206, 751-760. |
| [3] | Bhuckory S, Kays JC, Dennis AM (2019). In vivo biosensing using resonance energy transfer. Biosensors (Basel) 9, 76. |
| [4] | Bhupathi P, Elhassan A-Elgadir TM, Mohammed Ali RH, Sanaan Jabbar H, Gulnoza D, Joshi SK, Kadhem Abid M, Ahmed Said E, Alawadi A, Alsaalamy A (2023). Fluorescence resonance energy transfer (FRET)-based sensor for detection of foodborne pathogenic bacteria: a review. Crit Rev Anal Chem 1-18. [2023-11-02]. https://www.tandfonline.com/doi/full/10.1080/10408347.2023.2274050. |
| [5] |
Bogner M, Ludewig U (2007). Visualization of arginine influx into plant cells using a specific FRET-sensor. J Fluoresc 17, 350-360.
PMID |
| [6] | Bowler MW, Cliff MJ, Waltho JP, Blackburn GM (2010). Why did nature select phosphate for its dominant roles in biology? New J Chem 34, 784-794. |
| [7] |
Chaudhuri B, Hörmann F, Frommer WB (2011). Dynamic imaging of glucose flux impedance using FRET sensors in wild-type Arabidopsis plants. J Exp Bot 62, 2411-2417.
DOI PMID |
| [8] | Chaudhuri B, Hörmann F, Lalonde S, Brady SM, Orlando DA, Benfey P, Frommer WB (2008). Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J 56, 948-962. |
| [9] |
Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020). Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62, 25-54.
DOI |
| [10] | Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527-532. |
| [11] | Chen YN, Cartwright HN, Ho CH (2022). In vivo visualization of nitrate dynamics using a genetically encoded fluorescent biosensor. Sci Adv 8, eabq4915. |
| [12] | Davies PJ (2010). The plant hormones:their nature, occurrence, and functions. In: Davies PJ, ed. Plant Hormones: Biosynthesis, Signal Transduction, Action! 3rd edn. Dordrecht: Springer. pp. 1-15. |
| [13] | De Col V, Fuchs P, Nietzel T, Elsässer M, Voon CP, Candeo A, Seeliger I, Fricker MD, Grefen C, Møller IM, Bassi A, Lim BL, Zancani M, Meyer AJ, Costa A, Wagner S, Schwarzländer M (2017). ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 6, e26770. |
| [14] |
Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB (2006). Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18, 2314-2325.
DOI PMID |
| [15] | Ding NN, Zhou SH, Deng Y (2021). Transcription-factor- based biosensor engineering for applications in synthetic biology. ACS Synth Biol 10, 911-922. |
| [16] |
Fehr M, Frommer WB, Lalonde S (2002). Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc Natl Acad Sci USA 99, 9846-9851.
DOI PMID |
| [17] | Fehr M, Lalonde S, Lager I, Wolff MW, Frommer WB (2003). In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J Biol Chem 278, 19127-19133. |
| [18] | Forde BG, Lea PJ (2007). Glutamate in plants: metabolism, regulation, and signaling. J Exp Bot 58, 2339-2358. |
| [19] | Förster T (1965). Delocalized excitation and excitation transfer. In: SunanogluO,ed. Modern Quantum Chemistry. New York: Academic. |
| [20] |
Gao SP, Chu CC (2020). Gibberellin metabolism and signaling: targets for improving agronomic performance of crops. Plant Cell Physiol 61, 1902-1911.
DOI PMID |
| [21] |
Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276, 29188-29194.
DOI PMID |
| [22] | Gu H, Lalonde S, Okumoto S, Looger LL, Scharff- Poulsen AM, Grossman AR, Kossmann J, Jakobsen I, Frommer WB (2006). A novel analytical method for in vivo phosphate tracking. FEBS Lett 580, 5885-5893. |
| [23] |
Hamers D, van Voorst Vader L, Borst JW, Goedhart J (2014). Development of FRET biosensors for mammalian and plant systems. Protoplasma 251, 333-347.
DOI PMID |
| [24] | Herud-Sikimić O, Stiel AC, Kolb M, Shanmugaratnam S, Berendzen KW, Feldhaus C, Höcker B, Jürgens G (2021). A biosensor for the direct visualization of auxin. Nature 592, 768-772. |
| [25] | Ho CH, Frommer WB (2014). Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/ NRT1.1 and oligopeptide transporters. eLife 3, e01917. |
| [26] | Hu HT, Qian TT, Yang L (2022). Detection of reactive oxygen species using H2DCFDA probe in plant. Chin Bull Bot 57, 320-326. (in Chinese) |
|
胡海涛, 钱婷婷, 杨玲 (2022). 基于H2DCFDA荧光探针的植物活性氧检测方法. 植物学报 57, 320-326.
DOI |
|
| [27] |
Imamura H, Huynh Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H (2009). Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA 106, 15651-15656.
DOI PMID |
| [28] | Jones AM, Danielson JÅ, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3, e01741. |
| [29] | Jones AM, Grossmann G, Danielson JÅH, Sosso D, Chen LQ, Ho CH, Frommer WB (2013). In vivo biochemistry: applications for small molecule biosensors in plant biology. Curr Opin Plant Biol 16, 389-395. |
| [30] | Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012). FRET- based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69, 181-192. |
| [31] | Lakowicz JR (1999). Principles of Fluorescence Spectroscopy, 2nd edn. New York: Kluwer Academic/Plenum Publishers. pp. 11. |
| [32] |
Lam HK, McAdam SAM, McAdam EL, Ross JJ (2015). Evidence that chlorinated auxin is restricted to the Fabaceae but not to the Fabeae. Plant Physiol 168, 798-803.
DOI PMID |
| [33] | Lanquar V, Grossmann G, Vinkenborg JL, Merkx M, Thomine S, Frommer WB (2014). Dynamic imaging of cytosolic zinc in Arabidopsis roots combining FRET sensors and RootChip technology. New Phytol 202, 198-208. |
| [34] |
Laube B, Schemm R, Betz H (2004). Molecular determinants of ligand discrimination in the glutamate-binding pocket of the NMDA receptor. Neuropharmacology 47, 994-1007.
PMID |
| [35] | Ma GJ, Satheesh V, Lei MG (2022). Intracellular phosphate sensing in plants. Mol Plant 15, 1831-1833. |
| [36] |
Michener JK, Thodey K, Liang JC, Smolke CD (2012). Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng 14, 212-222.
DOI PMID |
| [37] | Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882-887. |
| [38] |
Monshausen GB (2012). Visualizing Ca2+ signatures in plants. Curr Opin Plant Biol 15, 677-682.
DOI PMID |
| [39] |
Mukherjee P, Banerjee S, Wheeler A, Ratliff LA, Irigoyen S, Garcia LR, Lockless SW, Versaw WK (2015). Live imaging of inorganic phosphate in plants with cellular and subcellular resolution. Plant Physiol 167, 628-638.
DOI PMID |
| [40] |
Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20, 87-90.
DOI PMID |
| [41] | Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A (2004). Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA 101, 10554-10559. |
| [42] |
O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016). Nitrate transport, sensing, and responses in plants. Mol Plant 9, 837-856.
DOI PMID |
| [43] |
Okumoto S, Pilot G (2011). Amino acid export in plants: a missing link in nitrogen cycling. Mol Plant 4, 453-463.
DOI PMID |
| [44] | Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM (2017). In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3, 803-813. |
| [45] | Rowe J, Grangé-Guermente M, Exposito-Rodriguez M, Wimalasekera R, Lenz MO, Shetty KN, Cutler SR, Jones AM (2023). Next-generation ABACUS biosensors reveal cellular ABA dynamics driving root growth at low aerial humidity. Nat Plants 9, 1103-1115. |
| [46] | Sami F, Siddiqui H, Hayat S (2019). Interaction of glucose and phytohormone signaling in plants. Plant Physiol Biochem 135, 119-126. |
| [47] | Shi SB, Ang EL, Zhao HM (2018). In vivo biosensors: mechanisms, development, and applications. J Ind Microbiol Biotechnol 45, 491-516. |
| [48] | Siddiqui H, Sami F, Hayat S (2020). Glucose: sweet or bitter effects in plants—a review on current and future perspective. Carbohydr Res 487, 107884. |
| [49] | Stanton C, Sanders D, Krämer U, Podar D (2022). Zinc in plants: integrating homeostasis and biofortification. Mol Plant 15, 65-85. |
| [50] |
Steinhorst L, He GF, Moore LK, Schültke S, Schmitz- Thom I, Cao YB, Hashimoto K, Andrés Z, Piepenburg K, Ragel P, Behera S, Almutairi BO, Batistič O, Wyganowski T, Köster P, Edel KH, Zhang CX, Krebs M, Jiang CF, Guo Y, Quintero FJ, Bock R, Kudla J (2022). A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev Cell 57, 2081-2094.
DOI PMID |
| [51] |
Sun YJ, Rose J, Wang BC, Hsiao CD (1998). The structure of glutamine-binding protein complexed with glutamine at 1.94 Å resolution: comparisons with other amino acid binding proteins. J Mol Biol 278, 219-229.
PMID |
| [52] | Swanson SJ, Gilroy S (2013). Imaging changes in cytoplasmic calcium using the Yellow Cameleon 3.6 biosensor and confocal microscopy. In: MunnikT, HeilmannI,eds. Plant Lipid Signaling Protocols. Totowa: Humana. pp. 291-302. |
| [53] |
Tegeder M, Rentsch D (2010). Uptake and partitioning of amino acids and peptides. Mol Plant 3, 997-1011.
DOI PMID |
| [54] |
Tian W, Wang C, Gao QF, Li LG, Luan S (2020). Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat Plants 6, 750-759.
DOI PMID |
| [55] |
Turner APF (2013). Biosensors: sense and sensibility. Chem Soc Rev 42, 3184-3196.
DOI PMID |
| [56] |
Uslu VV, Grossmann G (2016). The biosensor toolbox for plant developmental biology. Curr Opin Plant Biol 29, 138-147.
DOI PMID |
| [57] |
Evers TH, Dekkers LM, Meijer EW, Klomp LWJ, Merkx M (2007). Variation of linker length in ratiometric fluorescent sensor proteins allows rational tuning of Zn(II) affinity in the picomolar to femtomolar range. J Am Chem Soc 129, 3494-3495.
PMID |
| [58] | Verma AK, Noumani A, Yadav AK, Solanki PR (2023). FRET based biosensor: principle applications recent advances and challenges. Diagnostics (Basel) 13, 1375. |
| [59] |
Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M (2009). Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods 6, 737-740.
DOI PMID |
| [60] | Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014). FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3, e01739. |
| [61] |
Walia A, Waadt R, Jones AM (2018). Genetically encoded biosensors in plants: pathways to discovery. Annu Rev Plant Biol 69, 497-524.
DOI PMID |
| [62] | Wang XH, Feng CX, Tian LL, Hou CC, Tian W, Hu B, Zhang Q, Ren ZJ, Niu Q, Song JJ, Kong DD, Liu LY, He YK, Ma LG, Chu CC, Luan S, Li LG (2021). A transceptor-channel complex couples nitrate sensing to calcium signaling in Arabidopsis. Mol Plant 14, 774-786. |
| [63] | Wang YZ, Zhou YL, Liang JS (2022). Characterization of organellar-specific ABA responses during environmental stresses in tobacco cells and Arabidopsis plants. Cells 11, 2039. |
| [64] |
Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 464, 123-128.
DOI PMID |
| [65] | Wu YX, Jiang TY (2022). Developments in FRET- and BRET-based biosensors. Micromachines (Basel) 13, 1789. |
| [66] | Yang HY, Bogner M, Stierhof YD, Ludewig U (2010). H+-independent glutamine transport in plant root tips. PLoS One 5, e8917. |
| [67] | Yoshida T, Alfaqaan S, Sasaoka N, Imamura H (2017). Application of FRET-based biosensor “ATeam” for visualization of ATP levels in the mitochondrial matrix of living mammalian cells. In: MokranjacD, PerocchiF,eds. Mitochondria. New York: Humana Press. pp. 231-243. |
| [68] | Yoshinari A, Moe-Lange J, Kleist TJ, Cartwright HN, Quint DA, Ehrhardt DW, Frommer WB, Nakamura M (2021). Using genetically encoded fluorescent biosensors for quantitative in vivo imaging. In: Sanchez-SerranoJJ, SalinasJ, eds. Arabidopsis Protocols. New York: Humana Press. pp. 303-322. |
| [69] |
Zhang SS, Pan YJ, Tian W, Dong MQ, Zhu HF, Luan S, Li LG (2017). Arabidopsis CNGC14 mediates calcium influx required for tip growth in root hairs. Mol Plant 10, 1004-1006.
DOI PMID |
| [70] | Zhang XJ, Hu Y, Yang XT, Tang YY, Han SY, Kang A, Deng HS, Chi YM, Zhu D, Lu Y (2019). Förster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens Bioelectron 138, 111314. |
| [71] | Zhu QD, Wang L, Dong QL, Chang S, Wen KX, Jia SH, Chu ZL, Wang HM, Gao P, Zhao HP, Han SC, Wang YD (2017). FRET-based glucose imaging identifies glucose signaling in response to biotic and abiotic stresses in rice roots. J Plant Physiol 215, 65-72. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超. 植物细胞自噬基因的功能与转录调控机制[J]. 植物学报, 2021, 56(2): 201-217. |
| [3] | 谭艳秋,孙姝璟,董静云,徐伟,王玲玲,王永飞. 植物细胞质膜离子通道研究进展[J]. 植物学报, 2019, 54(1): 102-118. |
| [4] | 刘洋, 张静, 王秋玲, 侯岁稳. 植物细胞自噬研究进展[J]. 植物学报, 2018, 53(1): 5-16. |
| [5] | 王明红, 马来, 郑小江, 胡一兵. 植物微流芯片——一种实时定量监测生长发育的 高通量整合分析平台[J]. 植物学报, 2015, 50(5): 637-. |
| [6] | 李永成;陶文沂. 内生真菌培养液对东北红豆杉细胞生长及紫杉醇合成的影响[J]. 植物学报, 2008, 25(05): 552-558. |
| [7] | 王荣 焦群英 魏德强. 植物细胞的生物力学研究现状与进展[J]. 植物学报, 2005, 22(04): 478-485. |
| [8] | 袁晓凡 赵兵 王玉春. 稀土元素在药用植物细胞和组织培养中的应用[J]. 植物学报, 2005, 22(01): 115-120. |
| [9] | 何群;尤瑞麟. 应用Steedman's wax 切片法观察植物细胞微管骨架[J]. 植物学报, 2004, 21(05): 547-555. |
| [10] | 黄艳 赵德修 李佐虎. 利用生物反应器培养植物细胞的研究进展(Ⅱ)[J]. 植物学报, 2001, 18(06): 665-671. |
| [11] | 黄艳 赵德修 李佐虎. 植物细胞生物反应器培养的研究进展(I)[J]. 植物学报, 2001, 18(05): 567-570. |
| [12] | 杨弘远 周嫦. 植物实验生殖生物学与生殖工程:现在与未来[J]. 植物学报, 1989, 6(04): 193-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||