

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 285-287.DOI: 10.11983/CBB19060 cstr: 32102.14.CBB19060
• 热点评 • 下一篇
收稿日期:2019-03-31
接受日期:2019-04-02
出版日期:2019-05-01
发布日期:2019-05-20
通讯作者:
王二涛
Received:2019-03-31
Accepted:2019-04-02
Online:2019-05-01
Published:2019-05-20
Contact:
Ertao Wang
摘要: 根际微生物影响植物的生长及环境适应性。不同种属、不同种群的植物影响其环境微生物群落; 反之, 根际微生物也影响宿主植物生长发育与生态适应性。植物与根际微生物的互作现象及其机制, 是生命科学研究关注的热点, 也是农业微生物利用的关键问题。近期, 中国科学家在该领域取得了突破性进展。通过对不同籼稻(Oryza sativa subsp. indica)和粳稻(O. sativa subsp. japonica)品种的根际微生物组进行研究, 发现籼稻根际比粳稻根际富集更多参与氮代谢的微生物群落, 且该现象与硝酸盐转运蛋白基因NRT1.1B在籼粳之间的自然变异相关联。通过对籼稻接种籼稻根际特异富集的微生物群体可以提高前者对有机氮的利用, 促进其生长。该研究揭示了籼稻和粳稻根际微生物分化的分子基础, 展示了利用根际微生物提高水稻营养高效吸收的应用前景。
王孝林, 王二涛. 根际微生物促进水稻氮利用的机制. 植物学报, 2019, 54(3): 285-287.
Xiaolin Wang, Ertao Wang. NRT1.1B Connects Root Microbiota and Nitrogen Use in Rice. Chinese Bulletin of Botany, 2019, 54(3): 285-287.
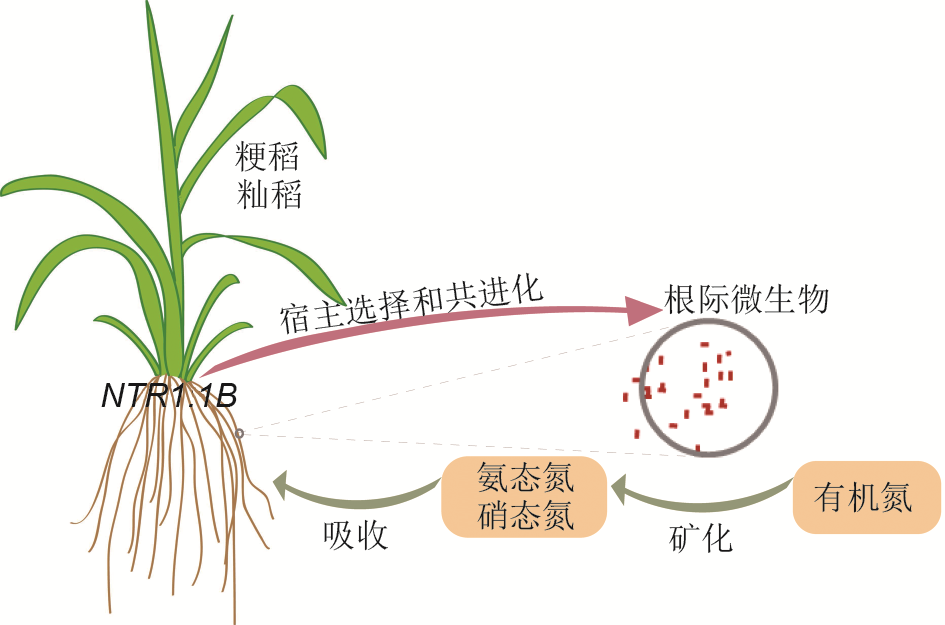
图1 水稻通过NRT1.1B基因协同根系微生物利用土壤氮元素籼稻比粳稻富集更多参与氮代谢的根系微生物。这些微生物将有机氮转化为氨态氮, 提高水稻氮元素利用效率。籼稻和粳稻特异富集的根系微生物与NRT1.1B基因相关联。
Figure 1 Rice plants coordinate root microbiota to utilize soil nitrogen by NRT1.1B Indica and japonica rice varieties recruit distinct root microb- iota. The biological progresses related to nitrogen metabolism are specifically enriched in indica-enriched bacteria. These bacteria transform organic nitrogen to ammonium, facilitating the nitrogen utilization in rice. NRT1.1B is associated with the recruitments of indica and japonica-specific bacterial taxa.
| [1] |
Beckers B, Op De Beeck M, Weyens N, Van Acker R, Van Montagu M, Boerjan W, Vangronsveld J ( 2016). Lignin engineering in field-grown poplar trees affects the endosphere bacterial microbiome. Proc Natl Acad Sci USA 113, 2312-2317.
DOI URL PMID |
| [2] |
Bender SF, Wagg C, van der Heijden MGA ( 2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31, 440-452.
DOI URL PMID |
| [3] |
Berendsen RL, Pieterse CM, Bakker PA ( 2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17, 478-486.
DOI URL PMID |
| [4] |
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P ( 2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91-95.
DOI URL PMID |
| [5] |
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P ( 2013). Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64, 807-838.
DOI URL PMID |
| [6] |
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM ( 2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fert Soils 48, 489-499.
DOI URL |
| [7] |
Duan L, Liu HB, Li XH, Xiao JH, Wang SP ( 2014). Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant 152, 486-500.
DOI URL PMID |
| [8] |
Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V ( 2015). Structure, variation, and assembly of the root- associated microbiomes of rice. Proc Natl Acad Sci USA 112, E911-E920.
DOI URL PMID |
| [9] |
Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC ( 2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47, 834-838.
DOI URL PMID |
| [10] |
Mbodj D, Effa-Effa B, Kane A, Manneh B, Gantet P, Laplaze L, Diedhiou AG, Grondin A ( 2018). Arbuscular mycorrhizal symbiosis in rice: establishment, environmental control and impact on plant growth and resistance to abiotic stresses. Rhizosphere 8, 12-26.
DOI URL |
| [11] |
Oldroyd GED, Murray JD, Poole PS, Downie JA ( 2011). The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45, 119-144.
DOI URL PMID |
| [12] |
Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT ( 2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA 112, E5013-E5020.
DOI URL PMID |
| [13] |
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET ( 2018). Core microbiomes for sustainable agroecosystems. Nat Plants 4, 247-257.
DOI URL |
| [14] |
Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, Gonzalez-Pena A, Peiffer J, Koren O, Shi Q, Knight R, Glavina del Rio T, Tringe SG, Buckler ES, Dangl JL, Ley RE ( 2018). Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc Natl Acad Sci USA 115, 7368-7373.
DOI URL |
| [15] | Zhang J, Liu Y, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J, Cao S, Wang X, Wang C, Wang H, Qu B, Fan GY, Yuan L, Garrido-Oter R, Chu C, Bai Y ( 2019). NRT1.1B contributes the association of root microbiota and nitrogen use in rice. Nat Biotechnol doi: https://doi.org/10.1038/s41587-019-0104-4. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [12] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [13] | 李柳, 刘庆华, 尹春英. 植物硒生物强化及微生物在其中的应用潜力[J]. 植物生态学报, 2023, 47(6): 756-769. |
| [14] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [15] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
