

植物学报 ›› 2016, Vol. 51 ›› Issue (1): 9-15.DOI: 10.11983/CBB14188 cstr: 32102.14.CBB14188
收稿日期:2014-10-27
接受日期:2015-03-11
出版日期:2016-01-01
发布日期:2016-02-01
通讯作者:
蒋甲福
作者简介:? 共同第一作者
基金资助:Yuying Qi, Yanli Zhan, Cuibo Wang, Fadi Chen, Jiafu Jiang*
Received:2014-10-27
Accepted:2015-03-11
Online:2016-01-01
Published:2016-02-01
Contact:
Jiang Jiafu
About author:? These authors contributed equally to this paper
摘要: RNA聚合酶II CTD磷酸化酶1 (CPL1)作为影响RNA聚合酶II磷酸化水平的重要因子, 在植物逆境响应、离子吸收以及成花诱导等生命过程中扮演重要角色。为深入探究CPL1参与植物开花时间调控的作用机制, 以拟南芥(Arabidopsis thaliana) AtCPL1突变体cpl1-3和fry2-1为研究材料, 观察了长日照条件下突变体与野生型的开花时间, 并利用荧光定量PCR技术对突变体中开花相关基因的表达情况进行了检测。结果表明: 在长日照条件下, 突变体cpl1-3和fry2-1抽薹时的莲座叶数目均显著多于野生型, 且表现出明显的开花时间延迟现象; 荧光定量PCR分析显示, 突变体cpl1-3和fry2-1中开花抑制因子miR156a、TOEs和SMZ基因的表达量较野生型显著升高, 而开花促进因子FT、FD、CO和miR172基因的表达量则显著降低。由此推测, AtCPL1通过调控miR156和miR172的表达水平进而影响下游开花相关基因TOEs、SMZ、FT、FD及CO等的表达, 从而实现对拟南芥开花时间的调控。
亓钰莹, 展妍丽, 王萃铂, 陈发棣, 蒋甲福. AtCPL1调控拟南芥开花的机制. 植物学报, 2016, 51(1): 9-15.
Yuying Qi, Yanli Zhan, Cuibo Wang, Fadi Chen, Jiafu Jiang. Mechanism of AtCPL1 in Regulating Flowering of Arabidopsis. Chinese Bulletin of Botany, 2016, 51(1): 9-15.
| Primer name | Sequence (5'-3') | Annotation |
|---|---|---|
| cpl1-3-F | TTGTCCTCATATTTGTGAGTTTTGG | |
| cpl1-3-R | CCTCAAATGAGCGCATGGTATT | |
| fry2-1- F | CTCAACTCAGTCTGACAGGATGCT | |
| fry2-1-R | TCCCAATACCCAAGTCAGTGATT | |
| UBQ10-F | GGCCTTGTATAATCCCTGATGAATAAG | Liu et al., 2011 |
| UBQ10-R | AAAGAGATAACAGGAACGGAAACATAGT | |
| FT-F | AGACGTTCTTGATCCGTTTA | Jung et al., 2007 |
| FT-R | GTAGATCTCAGCAAACTCGC | |
| CO-F | CCGGGTCTGCGAGTCATG | Yu et al., 2012 |
| CO-R | GGCATCATCTGCCTCACACA | |
| SOC1-F | GGTTTCTGAAGAAAATATGCAGCATTTG | Yang et al., 2010 |
| SOC1-R | GGCTACTCTCTTCATCACCTCTTCCAG | |
| FD-F | GCGCTAGGAAACAGGAATGCTTATACAAA | Yang et al., 2010 |
| FD-R | AGCTGTGGAAGACCGTTGAAGTGTG | |
| SMZ-F | AGGGAGAAGGAGCCATGAAGTTTGGTG | Yu et al., 2012 |
| SMZ-R | GTCTTCAGAGGTTTCATGGTTGCCATG | |
| miR156a-F | CTCAAGTTCATTGCCATTTTTAGG | Wang et al., 2009 |
| miR156a-R | GAGAGATTGAGACATAGAGAACGAAGA | |
| miR172a1-F | GGGTTTGAGTTTGAGTTTGA | Jung et al., 2007 |
| miR172a1-R | TGGATGGAATCCAAATAATC | |
| miR172a2-F | TCGGCGGATCCATGGAAGAAAGCTC | Jung et al., 2007 |
| miR172a2-R | TTTCTCAAGCTTTAGGTATTTGTAG | |
| miR172b1-F | CCGTCTTGAGTCTTGAAAAG | Jung et al., 2007 |
| miR172b1-R | GAAATACCTCCGATCTGTGA | |
| TOE1-F | CGAGTTATAATAATCCCGCCGAG | Yu et al., 2012 |
| TOE1-R | TTAAGGGTGTGGATAAAAGT | |
| TOE2-F | ATGGAGAACCACATGGCTGC | Yu et al., 2012 |
| TOE2-R | GGTGCTGTAGCTGCTACGGC | |
| TOE3-F | ACGAGGAACGGTCATAATCTTG | Yu et al., 2012 |
| TOE3-R | ATTCCCACCACTCGATTTCCT | |
| FLC-F | AGCCAAGAAGACCGAACTCA | Yu et al., 2012 |
| FLC-R | TTTGTCCAGCAGGTGACATC |
表1 引物序列
Table 1 Sequences of primers
| Primer name | Sequence (5'-3') | Annotation |
|---|---|---|
| cpl1-3-F | TTGTCCTCATATTTGTGAGTTTTGG | |
| cpl1-3-R | CCTCAAATGAGCGCATGGTATT | |
| fry2-1- F | CTCAACTCAGTCTGACAGGATGCT | |
| fry2-1-R | TCCCAATACCCAAGTCAGTGATT | |
| UBQ10-F | GGCCTTGTATAATCCCTGATGAATAAG | Liu et al., 2011 |
| UBQ10-R | AAAGAGATAACAGGAACGGAAACATAGT | |
| FT-F | AGACGTTCTTGATCCGTTTA | Jung et al., 2007 |
| FT-R | GTAGATCTCAGCAAACTCGC | |
| CO-F | CCGGGTCTGCGAGTCATG | Yu et al., 2012 |
| CO-R | GGCATCATCTGCCTCACACA | |
| SOC1-F | GGTTTCTGAAGAAAATATGCAGCATTTG | Yang et al., 2010 |
| SOC1-R | GGCTACTCTCTTCATCACCTCTTCCAG | |
| FD-F | GCGCTAGGAAACAGGAATGCTTATACAAA | Yang et al., 2010 |
| FD-R | AGCTGTGGAAGACCGTTGAAGTGTG | |
| SMZ-F | AGGGAGAAGGAGCCATGAAGTTTGGTG | Yu et al., 2012 |
| SMZ-R | GTCTTCAGAGGTTTCATGGTTGCCATG | |
| miR156a-F | CTCAAGTTCATTGCCATTTTTAGG | Wang et al., 2009 |
| miR156a-R | GAGAGATTGAGACATAGAGAACGAAGA | |
| miR172a1-F | GGGTTTGAGTTTGAGTTTGA | Jung et al., 2007 |
| miR172a1-R | TGGATGGAATCCAAATAATC | |
| miR172a2-F | TCGGCGGATCCATGGAAGAAAGCTC | Jung et al., 2007 |
| miR172a2-R | TTTCTCAAGCTTTAGGTATTTGTAG | |
| miR172b1-F | CCGTCTTGAGTCTTGAAAAG | Jung et al., 2007 |
| miR172b1-R | GAAATACCTCCGATCTGTGA | |
| TOE1-F | CGAGTTATAATAATCCCGCCGAG | Yu et al., 2012 |
| TOE1-R | TTAAGGGTGTGGATAAAAGT | |
| TOE2-F | ATGGAGAACCACATGGCTGC | Yu et al., 2012 |
| TOE2-R | GGTGCTGTAGCTGCTACGGC | |
| TOE3-F | ACGAGGAACGGTCATAATCTTG | Yu et al., 2012 |
| TOE3-R | ATTCCCACCACTCGATTTCCT | |
| FLC-F | AGCCAAGAAGACCGAACTCA | Yu et al., 2012 |
| FLC-R | TTTGTCCAGCAGGTGACATC |
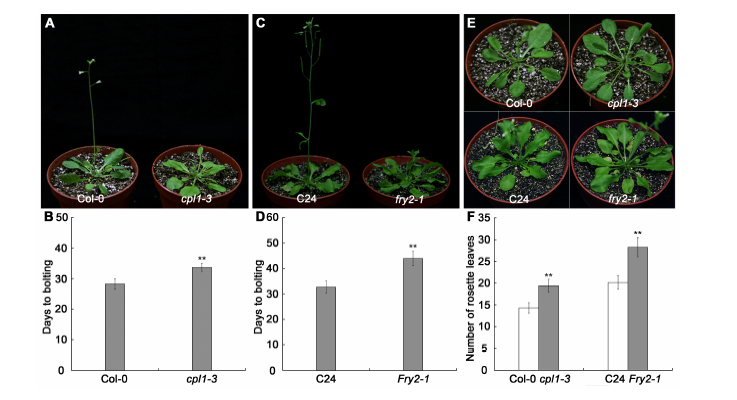
图1 长日照条件下cpl1-3和fry2-1突变体的开花表型 (A), (B) 突变体cpl1-3开花时间; (C), (D) 突变体fry2-1开花时间; (E), (F) cpl1-3和fry2-1突变体开花时的莲座叶数。数据用平均值±标准差表示; *和**分别表示在P≤0.05和P≤0.01水平上差异显著。
Figure 1 Phenotypic analysis of flowering of cpl1-3 and fry2-1 mutants under long days(A), (B) Flowering time of cpl1-3 mutant; (C), (D) Flowering time of fry2-1 mutant; (E), (F) The number of rosette leaves at flowering stage of cpl1-3 and fry2-1 mutants. Values are presented as means ± SD; * and ** represent significant differences at P≤0.05 and P≤0.01 level, respectively.

图2 长日照条件下cpl1-3 (A)和fry2-1 (B)突变体中开花相关基因的表达量 数据为平均值±标准差; *和**分别表示在P≤0.05和P≤0.01水平上差异显著。
Figure 2 Relative expression levels of the flowering related genes in cpl1-3 (A), fry2-1 (B) mutant under long daysValues are presented as means±SD; * and ** represent significant differences at P≤0.05 and P≤0.01 level, respectively.
| 1 | 韩玉波, 张飞雄 (2003). RNA聚合酶II中的CTD结构在mRNA转录和加工偶联过程中的重要作用. 遗传 25, 102-106. |
| 2 | 刘生财, 杨文文, 吴高杰, 赖钟雄 (2012). 高等植物成花的生理生化与分子机制研究进展. 亚热带农业研究 8, 37-41. |
| 3 | 孙昌辉, 邓晓建, 方军, 储成才 (2007). 高等植物开花诱导研究进展. 遗传 29, 1182-1190. |
| 4 | 徐妙云, 王磊 (2011). microRNA与植物花发育调控的研究进展. 中国农业科技导报 13, 9-16. |
| 5 | 许园园, 王燕, 柳李旺, 徐良, 王克磊, 张凤娇, 聂姗姗, 龚义勤 (2014). 萝卜抽薹开花促进因子RsFPF1基因克隆与功能分析. 南京农业大学学报 37, 31-38. |
| 6 | 袁秀云, 张仙云, 马杰, 秦喜庆 (2010). 植物开花的分子调控机理研究. 安徽农业科学 38, 614-616. |
| 7 | 张艺能, 周玉萍, 陈琼华, 黄小玲, 田长恩 (2014). 拟南芥开花时间调控的分子基础. 植物学报 49, 469-482. |
| 8 | Aksoy E, Jeong IS, Koiwa H (2013). Loss of function of Arabidopsis C-terminal domain phosphatase-like1 activates iron deficiency responses at the transcriptional level.Plant Physiol 161, 330-345. |
| 9 | Aukerman MJ, Sakai H (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2- like target genes.Plant Cell 15, 2730-2741. |
| 10 | Bang WY, Kim SW, Jeong IS, Koiwa H, Bahk JD (2008). The C-terminal region (640-967) of Arabidopsis CPL1 interacts with the abiotic stress- and ABA-responsive transcription factors.Biochem Bioph Res Co 372, 907-912. |
| 11 | Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis.Science 316, 1030-1033. |
| 12 | Duncan DB (1955). Multiple range and multiple T tests.Biometrics 11, 1-42. |
| 13 | Guan QM, Yue XL, Zeng HT, Zhu JH (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis.Plant Cell 26, 438-453. |
| 14 | Jeong IS, Aksoy E, Fukudome A, Akhter S, Hiraguri A, Fukuhara T, Bahk JD, Koiwa H (2013a). Arabidopsis C-terminal domain phosphatase-like 1 functions in miRNA accumulation and DNA methylation.PLoS One 8, e74739. |
| 15 | Jeong IS, Fukudome A, Aksoy E, Bang WY, Kim S, Guan QM, Bahk JD, May KA, Russell WK, Zhu JH, Koiwa H (2013b). Regulation of abiotic stress signaling by Arabidopsis C-terminal domain phosphatase-like 1 requires interaction with a K-homology domain-containing protein. PLoS One 8, e80509. |
| 16 | Jiang JF, Wang BS, Shen Y, Wang H, Feng Q, Shi HZ (2013). The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress- inducible gene expression.PLoS Genet 9, e1003625. |
| 17 | Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis.Plant Cell 19, 2736-2748. |
| 18 | Koiwa H, Barb AW, Xiong LM, Li F, McCully MG, Lee BH, Sokolchik I, Zhu JH, Gong ZZ, Reddy M, Sharkhuu A, Manabe Y, Yokoi S, Zhu JK, Bressan RA, Hasegawa PM (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development.Proc Natl Acad Sci USA 99,10893-10898. |
| 19 | Koiwa H, Hausmann S, Bang WY, Ueda A, Kondo N, Hiraguri A, Fukuhara T, Bahk JD, Yun DJ, Bressan RA, Hasegawa PM, Shuman S (2004). Arabidopsis C-ter- minal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases.Proc Natl Acad Sci USA 101, 14539-14544. |
| 20 | Liu TY, Aung K, Tseng CY, Chang TY, Chen YS, Chiou TJ (2011). Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis.Plant Physiol 156, 1176-1189. |
| 21 | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method.Methods 25, 402-408. |
| 22 | Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D (2012). Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1.Cell 151, 859-870. |
| 23 | Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M (2009). Repression of flowering by the miR172 target SMZ.PLoS Biol 7, e1000148. |
| 24 | Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz- Sommer Z, Yanofsky MF, Coupland G (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis.Science 288, 1613-1616. |
| 25 | Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis.Gene Dev 20, 898-912. |
| 26 | Wang JW, Czech B, Weigel D (2009). MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana.Cell 138, 738-749. |
| 27 | Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis.Cell 138, 750-759. |
| 28 | Xiong LM, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK (2002). Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis.Proc Natl Acad Sci USA 99, 10899-10904. |
| 29 | Yang WN, Jiang DH, Jiang JF, He YH (2010). A plant- specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis.Plant J 62, 663-673. |
| 30 | Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139, 770-778. |
| 31 | Yu S, Galvao VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BIN- DING-LIKE transcription factors.Plant Cell 24, 3320-3332. |
| 32 | Zhu QH, Helliwell CA (2011). Regulation of flowering time and floral patterning by miR172. J Exp Bot 62, 487-495. |
| [1] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 刘雨函, 曹启江, 张诗晗, 李益慧, 王菁, 谭晓萌, 刘筱儒, 王显玲. 拟南芥AtFTCD-L参与根系响应土壤紧实度的机制研究[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 景艳军, 林荣呈. 蓝光受体CRY2化身“暗黑舞者”[J]. 植物学报, 2024, 59(6): 878-882. |
| [4] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [5] | 陈艳晓, 李亚萍, 周晋军, 解丽霞, 彭永彬, 孙伟, 和亚男, 蒋聪慧, 王增兰, 郑崇珂, 谢先芝. 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024, 59(3): 481-494. |
| [6] | 杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础[J]. 植物学报, 2024, 59(3): 373-382. |
| [7] | 王钢, 王二涛. “卫青不败由天幸”——WeiTsing的广谱抗根肿病机理被揭示[J]. 植物学报, 2023, 58(3): 356-358. |
| [8] | 杨永青, 郭岩. 植物细胞质外体pH感受机制的解析[J]. 植物学报, 2022, 57(4): 409-411. |
| [9] | 支添添, 周舟, 韩成云, 任春梅. PAD4突变加速拟南芥酪氨酸降解缺陷突变体sscd1的程序性细胞死亡[J]. 植物学报, 2022, 57(3): 288-298. |
| [10] | 李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41. |
| [11] | 杨小凤, 李小蒙, 廖万金. 植物开花时间的遗传调控通路研究进展[J]. 生物多样性, 2021, 29(6): 825-842. |
| [12] | 张长生, 魏滔, 周玉萍, 范甜, 吕天晓, 田长恩. FLC调控植物成花的分子机制研究新进展[J]. 植物学报, 2021, 56(6): 651-663. |
| [13] | 车永梅, 孙艳君, 卢松冲, 侯丽霞, 范欣欣, 刘新. AtMYB77促进NO合成参与调控干旱胁迫下拟南芥侧根发育[J]. 植物学报, 2021, 56(4): 404-413. |
| [14] | 李秋信, 迟伟, 季代丽. CURT1调控类囊体膜弯曲的研究进展[J]. 植物学报, 2021, 56(4): 462-469. |
| [15] | 王婷, 羊欢欢, 赵弘巍, JosefVoglmeir, 刘丽. 蛋白质N-糖基化在拟南芥生长周期中的变化规律及去糖基化对根发育的影响[J]. 植物学报, 2021, 56(3): 262-274. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
