

植物学报 ›› 2021, Vol. 56 ›› Issue (6): 651-663.DOI: 10.11983/CBB21103 cstr: 32102.14.CBB21103
收稿日期:2021-06-29
接受日期:2021-09-10
出版日期:2021-11-01
发布日期:2021-11-12
通讯作者:
田长恩
作者简介:* E-mail: changentian@aliyun.com基金资助:
Changsheng Zhang, Tao Wei, Yuping Zhou, Tian Fan, Tianxiao Lü, Chang'en Tian( )
)
Received:2021-06-29
Accepted:2021-09-10
Online:2021-11-01
Published:2021-11-12
Contact:
Chang'en Tian
摘要: FLC是植物成花关键抑制因子, 主要通过结合到其下游2个关键的成花促进基因(FT和SOC1)启动子上而抑制二者的表达。此外, 还可以与其它调控因子结合调控开花。然而, 关于FLC在成花调控中的具体分子机制仍需深入研究。该文主要结合8条成花调控遗传途径, 梳理近年来与FLC相关的新进展, 并展望了未来的研究方向。
张长生, 魏滔, 周玉萍, 范甜, 吕天晓, 田长恩. FLC调控植物成花的分子机制研究新进展. 植物学报, 2021, 56(6): 651-663.
Changsheng Zhang, Tao Wei, Yuping Zhou, Tian Fan, Tianxiao Lü, Chang'en Tian. Progress in Flowering Regulation Mechanisms of FLC. Chinese Bulletin of Botany, 2021, 56(6): 651-663.
| 调控类型 | 调控因子 | 分子机制 | 参考文献 |
|---|---|---|---|
| 染色质重塑 | EFS | 一个组蛋白甲基转移酶, 通过下调FLC染色质H3Lys 4三甲基化水平, 抑制FLC表达; 同时也能与FRI形成复合物, 通过甲基化修饰促进FLC表达 | Kim et al., |
| LHP1 | 通过介导PRC2向染色质招募组蛋白甲基转移酶促进FLC染色质的H3K27me3积累, 进而维持FLC沉默 | Zhou et al., | |
| ICU11 | 一个2-氧戊二酸依赖的双加氧酶, 与非特异性组蛋白去甲基化有关, 其功能缺失后破坏低温诱导的PRC2复合物介导的FLC沉默, 从而导致晚花 | Bloomer et al., | |
| VRN1与VRN2 | 分别通过提高FLC启动子和第1个内含子区域内H3K9me2和H3K27 me2的水平, 抑制FLC表达 | Sung and Amasino, | |
| SWP与CZS | 二者形成复合体, 通过调控FLC染色质的H3K9和H3K27甲基化和H4去乙酰化, 抑制FLC表达 | Jiang et al., | |
| REF6 | 一个H3K27去甲基化转移酶, 使FLC去甲基化而抑制FLC表达 | Lu et al., | |
| PRMT5 | 通过调控FLC染色质H4R3对称性双甲基化, 下调FLC表达 | 牛丽芳, | |
| JMJ18 | 一个组蛋白H3K4去甲基化酶, 通过与FLC结合, 降低H3K4甲基化水平, 抑制FLC表达 | Yang et al., | |
| JMJ27 | 通过调控FLC染色质上的H3K9me2促进FLC表达 | Dutta et al., | |
| JMJ30与JMJ32 | JMJ30与JMJ32能够在高温下调节FLC位点的H3K27me3水平, 延缓H3K27me3介导的FLC抑制 | Bastow et al., | |
| PAF1复合物 | 具有RNA聚合酶II活性, 通过招募组蛋白甲基转移酶提高FLC染色质上H3K4和H3K36的甲基化水平, 从而促进FLC表达 | He et al., | |
| FRI | 通过上调FLC染色质H3K4三甲基化水平, 促进FLC表达 | He and Amasino, | |
| VIN3 | 通过参与FLC启动子区域组蛋白H3K9和H3K14的去乙酰化下调FLC表达 | Sung and Amasino, | |
| VIL1与VIL3 | 通过形成二聚体参与FLC的去乙酰化和甲基化, 进而抑制其表达 | Sung et al., | |
| FLD | 一个组蛋白乙酰化酶复合体, 对FLC组蛋白去乙酰化, 抑制FLC表达 | He et al., | |
| FVE | 对FLC染色质去乙酰化修饰, 抑制FLC表达 | Baek et al., | |
| HDA6 | 分别与FLD和HDA5互作, 形成复合物, 行使去乙酰化酶功能 | Cheng et al., | |
| HAM | 通过FLC及MAF3/MAF4染色质H4K5的乙酰化修饰抑制FLC表达 | Xiao et al., | |
| COOLAIR | 通过反义剪切调控FLC染色质上甲基化水平, 抑制FLC表达 | Csorba et al., | |
| 调控类型 | 调控因子 | 分子机制 | 参考文献 |
| 启动子结合 | SUF4 | 与FRI形成复合物后结合到FLC启动子, 抑制其表达 | Choi et al., |
| FLX与FLX4 | 与FRI形成复合物后结合到FLC启动子, 促进其表达 | Ding et al., | |
| PRC2复合物 | 通过其特异组分PHD抑制FLC转录起始蛋白与FLC启动子的结合, 引起FLC沉默 | Turck et al., | |
| TAF15b | 与FLC和COOLAIR的转录起始位点结合, 抑制FLC表达 | Eom et al., | |
| ABI4 | 与FLC的启动子CCAC基序结合, 促进FLC表达 | Shu et al., | |
| ABI5 | 与FLC启动子的ABRE/G-box (CACGTG)结合, 促进FLC的表达 | Xiong et al., | |
| 内含子及其它区域结合 | BIM | 结合到FLC的第1个内含子中的BR反应元件, 并招募去甲基化酶抑制H3K27三甲基化, 从而拮抗PRC2介导的FLC沉默 | Li et al., |
| BZR1 | 结合到FLC第1个内含子中的BR反应元件, 并招募去甲基化酶, 促进FLC表达 | Li et al., | |
| ASL | 其转录本结合到FLC的第1个内含子中, 影响其中关键位点的H3K27 me3水平 | Shin and Chekanova, | |
| JMJ28 | 靶向FLC的内含子区域和3′区域, 与FLC结合, 其是否通过调控FLC参与成花尚不确定 | Hung et al., | |
| mRNA结合 | FCA | 通过调控非编码RNA对邻近多聚腺苷化位点的选择, 从而在表观遗传学水平上调控FLC的表达 | Liu et al., |
| FY | 3′末端加工因子, 与FCA互作, 共同抑制FLC表达 | Simpson et al., | |
| GRP7与GRP8 | 富含甘氨酸的RNA结合蛋白, 二者都抑制FLC的表达, 具体分子机制有待阐明 | Wu et al., | |
| FPA | 具有RNA识别基序的RNA结合域, 参与FCA对RAN的修饰加工, 通过控制IncRNA的选择性聚腺苷化和FLC的3′端形成抑制FLC表达 | Cheng et al., | |
| 蛋白质稳定 | SIZ1 | 一个E3泛素连接酶, 能稳定FLC不被降解 | Son et al., |
表1 FLC基因表达的调控机制
Table1 Regulatory mechanism of FLC expression
| 调控类型 | 调控因子 | 分子机制 | 参考文献 |
|---|---|---|---|
| 染色质重塑 | EFS | 一个组蛋白甲基转移酶, 通过下调FLC染色质H3Lys 4三甲基化水平, 抑制FLC表达; 同时也能与FRI形成复合物, 通过甲基化修饰促进FLC表达 | Kim et al., |
| LHP1 | 通过介导PRC2向染色质招募组蛋白甲基转移酶促进FLC染色质的H3K27me3积累, 进而维持FLC沉默 | Zhou et al., | |
| ICU11 | 一个2-氧戊二酸依赖的双加氧酶, 与非特异性组蛋白去甲基化有关, 其功能缺失后破坏低温诱导的PRC2复合物介导的FLC沉默, 从而导致晚花 | Bloomer et al., | |
| VRN1与VRN2 | 分别通过提高FLC启动子和第1个内含子区域内H3K9me2和H3K27 me2的水平, 抑制FLC表达 | Sung and Amasino, | |
| SWP与CZS | 二者形成复合体, 通过调控FLC染色质的H3K9和H3K27甲基化和H4去乙酰化, 抑制FLC表达 | Jiang et al., | |
| REF6 | 一个H3K27去甲基化转移酶, 使FLC去甲基化而抑制FLC表达 | Lu et al., | |
| PRMT5 | 通过调控FLC染色质H4R3对称性双甲基化, 下调FLC表达 | 牛丽芳, | |
| JMJ18 | 一个组蛋白H3K4去甲基化酶, 通过与FLC结合, 降低H3K4甲基化水平, 抑制FLC表达 | Yang et al., | |
| JMJ27 | 通过调控FLC染色质上的H3K9me2促进FLC表达 | Dutta et al., | |
| JMJ30与JMJ32 | JMJ30与JMJ32能够在高温下调节FLC位点的H3K27me3水平, 延缓H3K27me3介导的FLC抑制 | Bastow et al., | |
| PAF1复合物 | 具有RNA聚合酶II活性, 通过招募组蛋白甲基转移酶提高FLC染色质上H3K4和H3K36的甲基化水平, 从而促进FLC表达 | He et al., | |
| FRI | 通过上调FLC染色质H3K4三甲基化水平, 促进FLC表达 | He and Amasino, | |
| VIN3 | 通过参与FLC启动子区域组蛋白H3K9和H3K14的去乙酰化下调FLC表达 | Sung and Amasino, | |
| VIL1与VIL3 | 通过形成二聚体参与FLC的去乙酰化和甲基化, 进而抑制其表达 | Sung et al., | |
| FLD | 一个组蛋白乙酰化酶复合体, 对FLC组蛋白去乙酰化, 抑制FLC表达 | He et al., | |
| FVE | 对FLC染色质去乙酰化修饰, 抑制FLC表达 | Baek et al., | |
| HDA6 | 分别与FLD和HDA5互作, 形成复合物, 行使去乙酰化酶功能 | Cheng et al., | |
| HAM | 通过FLC及MAF3/MAF4染色质H4K5的乙酰化修饰抑制FLC表达 | Xiao et al., | |
| COOLAIR | 通过反义剪切调控FLC染色质上甲基化水平, 抑制FLC表达 | Csorba et al., | |
| 调控类型 | 调控因子 | 分子机制 | 参考文献 |
| 启动子结合 | SUF4 | 与FRI形成复合物后结合到FLC启动子, 抑制其表达 | Choi et al., |
| FLX与FLX4 | 与FRI形成复合物后结合到FLC启动子, 促进其表达 | Ding et al., | |
| PRC2复合物 | 通过其特异组分PHD抑制FLC转录起始蛋白与FLC启动子的结合, 引起FLC沉默 | Turck et al., | |
| TAF15b | 与FLC和COOLAIR的转录起始位点结合, 抑制FLC表达 | Eom et al., | |
| ABI4 | 与FLC的启动子CCAC基序结合, 促进FLC表达 | Shu et al., | |
| ABI5 | 与FLC启动子的ABRE/G-box (CACGTG)结合, 促进FLC的表达 | Xiong et al., | |
| 内含子及其它区域结合 | BIM | 结合到FLC的第1个内含子中的BR反应元件, 并招募去甲基化酶抑制H3K27三甲基化, 从而拮抗PRC2介导的FLC沉默 | Li et al., |
| BZR1 | 结合到FLC第1个内含子中的BR反应元件, 并招募去甲基化酶, 促进FLC表达 | Li et al., | |
| ASL | 其转录本结合到FLC的第1个内含子中, 影响其中关键位点的H3K27 me3水平 | Shin and Chekanova, | |
| JMJ28 | 靶向FLC的内含子区域和3′区域, 与FLC结合, 其是否通过调控FLC参与成花尚不确定 | Hung et al., | |
| mRNA结合 | FCA | 通过调控非编码RNA对邻近多聚腺苷化位点的选择, 从而在表观遗传学水平上调控FLC的表达 | Liu et al., |
| FY | 3′末端加工因子, 与FCA互作, 共同抑制FLC表达 | Simpson et al., | |
| GRP7与GRP8 | 富含甘氨酸的RNA结合蛋白, 二者都抑制FLC的表达, 具体分子机制有待阐明 | Wu et al., | |
| FPA | 具有RNA识别基序的RNA结合域, 参与FCA对RAN的修饰加工, 通过控制IncRNA的选择性聚腺苷化和FLC的3′端形成抑制FLC表达 | Cheng et al., | |
| 蛋白质稳定 | SIZ1 | 一个E3泛素连接酶, 能稳定FLC不被降解 | Son et al., |
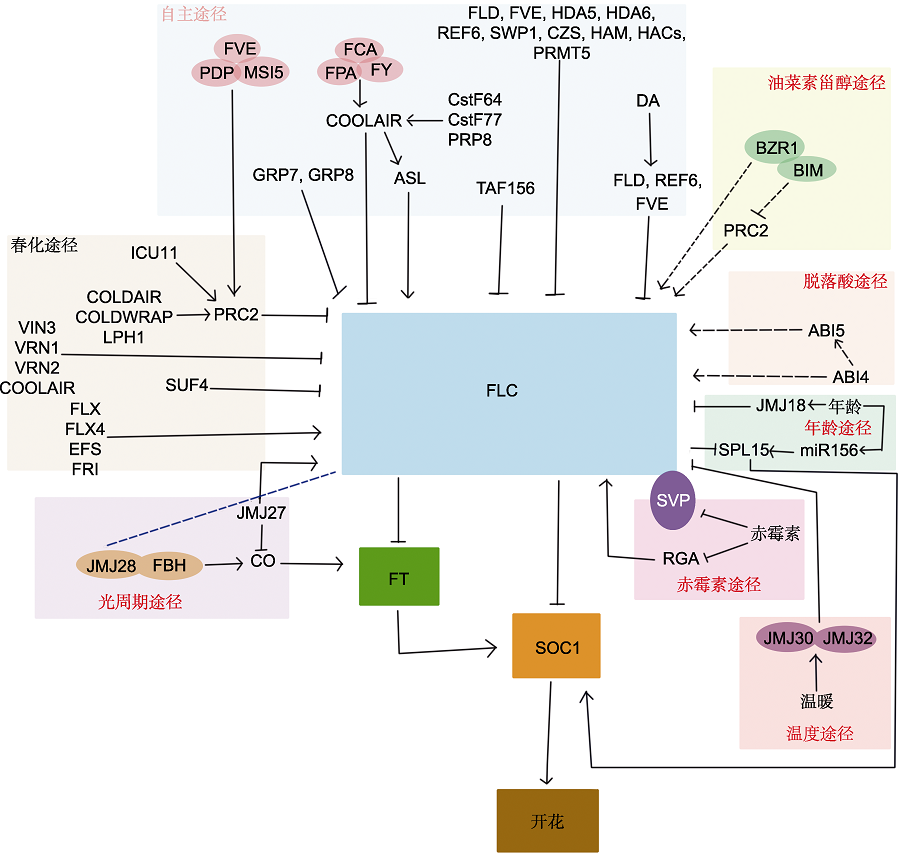
图1 FLC与成花调控网络 图中箭头表示促进, 钝线表示抑制。蓝色虚线表示有待研究。由于目前油菜素甾醇(BR)途径和脱落酸(ABA)途径证据较少, 故用虚线箭头表示。(1) 自主途径中, FVE、PDP和MSI5共同调控PRC2复合物, 抑制FLC表达; COOLAIR受FCA、FY和FPA影响, 同时, CstF64、CstF77和PRP8反义剪切产生的ASL维持FLC的甲基化修饰; FY与FCA互作, 在染色质水平上抑制FLC表达; FPA与FCA互作, 控制IncRNA的选择性聚腺苷化和FLC的3′端形成; FLD编码组蛋白乙酰化酶复合体, 抑制FLC表达; FVE下调FLC表达; FVE与FLD互作, 两者均参与组蛋白去乙酰化复合物, 但其机制尚不清楚; 组蛋白去乙酰化酶HDA5与HDA6互作, 两者均显示去乙酰化酶活性; HDA6与FLD互作, 这4种蛋白(FVE、FLD、HDA5和HDA6)可能形成蛋白复合物, 并通过组蛋白修饰相互作用, 抑制FLC表达; SWP1和CZS能形成SWP1/CZS复合体, 通过甲基化和去乙酰化调控FLC; REF6调控FLC的H3K27去甲基化; HAM参与FLC及MAF3/MAF4染色质H4K5的乙酰化修饰; HAC可能通过翻译后修饰FPA蛋白间接影响FLC表达; PRMT5介导FLC染色质H4R3对称性双甲基化,下调FLC表达; DA促进FLD、REF6和FVE的转录, 抑制FLC表达; GRP7的突变体中FLC表达上调并表现延迟开花, GRP8还能进一步晚花, 二者都能抑制FLC表达。(2) 年龄途径中, JMJ18直接抑制FLC的表达; SPL15作为FLC下游的靶标, FLC直接结合SPL15的启动子下调其表达。(3) 春化途径FRI复合物中的SUF4通过转录抑制FLC的表达, EFS能激活转录因子FLX和FLX4, 促进FLC的表达。(4) 光周期途径JMJ28和FBH共同调控CO, 促进FT表达, JMJ28与FLC结合, 但是否调控FLC表达还有待研究; JMJ27在光周期途径中通过抑制CO表达和促进FLC表达来参与成花调控。(5) 温度途径中, JMJ30/JMJ32在温暖条件下能够抑制FLC表达, 进而解除FLC对FT的抑制。(6) 赤霉素(GA)途径中, DEELA蛋白与FLC之间存在互作, DEELA蛋白中的RGA能直接促进FLC对SOC1的抑制; 同时SVP-FLC复合物也能抑制SOC1的表达。(7) BR通路中, BZR1能够与FLC内含子中的BR元件结合, 调控FLC表达, 同时BZR1和相互作用的MYC-like蛋白(BIM)与FLC第1个内含子中的BR反应元件结合, 对PRC2介导的FLC染色质沉默起拮抗作用。(8) ABA通路中, ABI4和ABI5既能通过启动子结合促进FLC表达, 也能共同作用促进FLC表达。
Figure 1 Schematic diagram of the FLC in flowering time regulation Arrows represent gene activation and blunted lines represent gene repression. The blue dotted line indicates the pathways that are not identified. The brassinosteroids (BR) pathway and abscisic acid (ABA) pathway are represented by dotted lines because there is less evidence. (1) Regulation of the PRC2 complex by FVE, PDP, and MSI5 in the autonomous pathway in repressing FLC; COOLAIR was affected by FCA, FY, FPA, while ASL generated by antisense splicing at Cstf64, Cstf77, and PRP8 maintained the methylation modification of FLC; FY interacts with FCA to repress FLC expression at the chromatin level; FPA interacts with FCA to control selective polyadenylation of IncRNA and 3′ end formation of FLC; FLD encodes a histone acetylase complex that represses FLC expression; FVE downregulates FLC expression; FVE interacts with FLD, both of which participate in the histone deacetylation complexes, which mechanism is currently unknown; the histone deacetylase HDA5 interacts with HDA6 and both display deacetylase activity; HDA6 interacts with FLD, and these 4 proteins (FVE, FLD, HDA5 and HDA6) may exist in protein complexes and repress FLC expression through histone modification interactions; SWP1 and CZS form the complex to regulate FLC through methylation and deacetylation; REF6 regulates H3K27 demethylation at FLC; HAM is involved in acetylation of H4K5 on chromatin at FLC and MAF3/MAF4; HAC may affect FLC expression indirectly through posttranslational modification of FPA proteins; PRMT5 mediates FLC chromatin H4R3 symmetric dimethylation, downregulating FLC expression; DA promoted the transcription of FLD, REF6 and FVE and inhibited FLC expression; in the mutants of GRP7, the expression of FLC was up-regulated and delayed flowering, and GRP8 could further late flowering, both of which could inhibit the expression of FLC. (2) The expression of FLC is directly inhibited by JMJ18 in the age pathway; SPL15 is a downstream target of FLC, FLC directly bind to the promoter of SPL15 to downregulate its expression. (3) SUF4 in the FRI complex of the vernalization pathway transcriptionally represses FLC expression, while EFS can activate the transcription factors FLX and FLX4 to promote FLC expression. (4) In the photoperiod pathway, JMJ28 and FBH jointly regulate CO, promote FT expression, and bind JMJ28 to FLC, but whether it regulates the expression of FLC remains to be investigated; JMJ27 functions in the photoperiod pathway to participate in the flowering regulation by repressing CO expression and promoting FLC expression. (5) JMJ30/JMJ32 in the temperature pathway is able to repress FLC expression under warm conditions and release the expression of FT. (6) An interaction between DEELA proteins and FLC was found in the gibberellins (GA) pathway, and RGA in DEELA proteins can directly promote the inhibition of SOC1 by FLC; at the same time, the SVP-FLC complex can also inhibit the expression of SOC1. (7) BZR1 in the BR pathway is able to bind BR elements in the FLC intron and regulate FLC expression. Meanwhile, BZR1 interacts with MYC-like protein (BIM) and binds to BR responsive elements in the first intron of FLC, which plays an antagonistic role in the PRC2-mediated silencing of FLC. (8) In the ABA pathway, ABI4 and ABI5 can not only promote the expression of FLC through promoter binding, but also promote the expression of FLC together.
| [1] |
陈唯, 曾晓贤, 谢楚萍, 田长恩, 周玉萍 (2019). 植物内源ABA水平的动态调控机制. 植物学报 54, 677-687.
DOI |
| [2] | 李勇, 胡宗利, 顾峰, 胡功铃, 陈国平 (2009). 拟南芥春化作用相关基因FLC的表达调控及天然早花突变体的研究进展. 生命科学 21, 335-342. |
| [3] | 刘娟, 黎黎, 陆柄辰, 邓朴, 艾辛 (2020). 温度调控植物开花研究进展. 应用与环境生物学报 26, 713-721. |
| [4] | 牛丽芳 (2009). 蛋白精氨酸甲基转移酶AtPRMT调控拟南芥开花时间的分子机理研究. 博士论文. 北京: 中国科学院遗传与发育生物学研究所. pp. 1-148. |
| [5] | 夏志强, 何奕昆, 鲍时来, 种康 (2007). 植物开花的组蛋白甲基化调控分子机理. 植物学通报 24, 275-283. |
| [6] | 肖旭峰, 范淑英 (2013). 抽薹开花抑制因子FLC表观遗传调控研究进展. 中国蔬菜 (22), 1-8. |
| [7] | 张艺能, 周玉萍, 陈琼华, 黄小玲, 田长恩 (2014). 拟南芥开花时间调控的分子基础. 植物学报 49, 469-482. |
| [8] | Baek IS, Park HY, You MK, Lee JH, Kim JK (2008). Functional conservation and divergence of FVE genes that control flowering time and cold response in rice and Arabidopsis. Mol Cells 26, 368-372. |
| [9] |
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164-167.
DOI URL |
| [10] |
Berry S, Dean C (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83, 133-148.
DOI URL |
| [11] | Bloomer RH, Hutchison CE, Bäurle I, Walker J, Fang XF, Perera P, Velanis CN, Gümüs S, Spanos C, Rappsilber J, Feng XQ, Goodrich J, Dean C (2020). The Arabidopsis epigenetic regulator ICU11 as an accessory protein of polycomb repressive complex 2. Proc Natl Acad Sci USA 117, 16660-16666. |
| [12] |
Bulgakov VP, Avramenko TV (2020). Linking brassinosteroid and ABA signaling in the context of stress acclimation. Int J Mol Sci 21, 5108.
DOI URL |
| [13] |
Cheng JZ, Zhou YP, Lv TX, Xie CP, Tian CE (2017). Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol Mol Biol Plants 23, 477-485.
DOI URL |
| [14] |
Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289-303.
DOI URL |
| [15] |
Chowdhury Z, Mohanty D, Giri MK, Venables BJ, Chaturvedi R, Chao A, Petros RA, Shah J (2020). Dehydroabietinal promotes flowering time and plant defense in Arabidopsis via the autonomous pathway genes FLOWERING LOCUS D, FVE, and RELATIVE OF EARLY FLOWERING 6. J Exp Bot 71, 4903-4913.
DOI URL |
| [16] |
Corbesier L, Vincent C, Jang S, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030-1033.
PMID |
| [17] |
Csorba T, Questa JI, Sun QW, Dean C (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111, 16160-16165.
DOI URL |
| [18] |
de Folter S, Angenent GC (2006). trans meets cis in MADS science. Trends Plant Sci 11, 224-231.
DOI URL |
| [19] |
Deng WW, Liu CY, Pei YX, Deng X, Niu LF, Cao XF (2007). Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol 143, 1660-1668.
DOI URL |
| [20] |
Deng WW, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108, 6680-6685.
DOI URL |
| [21] |
Ding L, Kim SY, Michaels SD (2013). FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol 163, 243-252.
DOI PMID |
| [22] |
Dutta A, Choudhary P, Caruana J, Raina R (2017). JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J 91, 1015-1028.
DOI URL |
| [23] |
Eom H, Park SJ, Kim MK, Kim H, Kang H, Lee I (2018). TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J 93, 79-91.
DOI URL |
| [24] |
Fang XF, Wu Z, Raitskin O, Webb K, Voigt P, Lu T, Howard M, Dean C (2020). The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc Natl Acad Sci USA 117, 15316-15321.
DOI URL |
| [25] | Gan ES, Xu YF, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T (2014). Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5, 5098. |
| [26] |
Greb T, Mylne JS, Crevillen P, Geraldo N, An HL, Gendall AR, Dean C (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17, 73-78.
DOI URL |
| [27] |
Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B (2010). ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol 153, 1062-1073.
DOI PMID |
| [28] |
He YH (2009). Control of the transition to flowering by chromatin modifications. Mol Plant 2, 554-564.
DOI URL |
| [29] |
He YH (2012). Chromatin regulation of flowering. Trends Plant Sci 17, 556-562.
DOI URL |
| [30] |
He YH, Amasino RM (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci 10, 30-35.
DOI URL |
| [31] |
He YH, Doyle MR, Amasino RM (2004). PAF1-complex- mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18, 2774-2784.
DOI URL |
| [32] |
He YH, Michaels SD, Amasino RM (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751-1754.
DOI URL |
| [33] |
Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46, 183-192.
PMID |
| [34] |
Heo JB, Sung S (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76-79.
DOI URL |
| [35] |
Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG (2010a). Alternative polyadenylation of antisense RNAs and flowering time control. Biochem Soc Trans 38, 1077-1081.
DOI URL |
| [36] |
Hornyik C, Terzi LC, Simpson GG (2010b). The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18, 203-213.
DOI URL |
| [37] |
Hung FY, Lai YC, Wang JH, Feng YR, Shih YH, Chen JH, Sun HC, Yang SG, Li CL, Wu KQ (2021). The Arabidopsis histone demethylase JMJ28 regulates CONSTANS by interacting with FBH transcription factors. Plant Cell 33, 1196-1211.
DOI URL |
| [38] |
Ietswaart R, Wu Z, Dean C (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet 28, 445-453.
DOI PMID |
| [39] |
Jiang DH, Yang WN, He YH, Amasino RM (2007). Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975-2987.
DOI URL |
| [40] | Jung JH, Park JH, Lee S, To KT, Kim JM, Seki M, Park CM (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25, 4378-4390. |
| [41] |
Kennedy A, Geuten K (2020). The role of FLOWERING LOCUS C relatives in cereals. Front Plant Sci 11, 617340.
DOI URL |
| [42] |
Kim DH, Sung S (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell 40, 302-312.
DOI URL |
| [43] |
Kim SY, He YH, Jacob Y, Noh YS, Michaels S, Amasino R (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17, 3301-3310.
DOI URL |
| [44] | Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR (2012). EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 8, e100 2512. |
| [45] |
Li D, Liu C, Shen LS, Wu Y, Chen HY, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15, 110-120.
DOI URL |
| [46] |
Li M, An F, Li W, Ma M, Feng Y, Zhang X, Guo H (2016). DELLA proteins interact with FLC to repress flowering transition. J Integr Plant Biol 58, 642-655.
DOI URL |
| [47] |
Li ZC, Jiang DH, He YH (2018a). FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plant 4, 836-846.
DOI URL |
| [48] |
Li ZC, Ou Y, Zhang ZC, Li JM, He YH (2018b). Brassinosteroid signaling recruits histone 3 lysine-27 demethylation activity to FLOWERING LOCUS C chromatin to inhibit the floral transition in Arabidopsis. Mol Plant 11, 1135-1146.
DOI URL |
| [49] |
Liu CY, Lu FL, Cui X, Cao XF (2010a). Histone methylation in higher plants. Annu Rev Plant Biol 61, 395-420.
DOI URL |
| [50] |
Liu FQ, Marquardt S, Lister C, Swiezewski S, Dean C (2010b). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94-97.
DOI URL |
| [51] |
Liu FQ, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28, 398-407.
DOI URL |
| [52] |
Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF (2011a). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43, 715-719.
DOI URL |
| [53] |
Lu FL, Li GL, Cui X, Liu CY, Wang XJ, Cao XF (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50, 886-896.
DOI URL |
| [54] |
Lu SX, Knowles SM, Webb CJ, Celaya BR, Cha C, Siu JP, Tobin EM (2011b). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol 155, 906-915.
DOI URL |
| [55] |
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737-745.
PMID |
| [56] |
Marquardt S, Raitskin O, Wu Z, Liu FQ, Sun QW, Dean C (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54, 156-165.
DOI URL |
| [57] |
Michaels SD (2009). Flowering time regulation produces much fruit. Curr Opin Plant Biol 12, 75-80.
DOI PMID |
| [58] |
Michaels SD, Amasino RM (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949-956.
PMID |
| [59] |
Monteagudo A, Igartua E, Contreras-Moreira B, Gracia MP, Ramos J, Karsai I, Casas AM (2019). Fine-tuning of the flowering time control in winter barley: the importance of HvOS2 and HvVRN2 in non-inductive conditions. BMC Plant Biol 19, 113.
DOI PMID |
| [60] |
Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004). Divergent roles of a pair of homologous Jumonji/Zinc-Finger-Class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601-2613.
DOI URL |
| [61] | Pajoro A, Severing E, Angenent GC, Immink RGH (2017). Histone H3 lysine 36 methylation affects temperature- induced alternative splicing and flowering in plants. Genome Biol 18, 102. |
| [62] |
Park BS, Sang WG, Yeu SY, Choi YD, Paek NC, Kim MC, Song JT, Seo HS (2007). Post-translational regulation of FLC is mediated by an E3 ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci 173, 269-275.
DOI URL |
| [63] |
Qi HD, Lin Y, Ren QP, Wang YY, Xiong F, Wang XL (2019). RNA Splicing of FLC modulates the transition to flowering. Front Plant Sci 10, 1625.
DOI URL |
| [64] |
Qin F, Yu BZ, Li WQ (2021). Heat shock protein 101 (HSP101) promotes flowering under nonstress conditions. Plant Physiol 186, 407-419.
DOI URL |
| [65] |
Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G (2019). Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET of FLC and SVP1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet 15, e1008065.
DOI URL |
| [66] |
Rosa S, Duncan S, Dean C (2016). Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun 7, 13031.
DOI URL |
| [67] |
Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS- box genes. Nat Commun 4, 2280.
DOI PMID |
| [68] |
Sasaki E, Frommlet F, Nordborg M (2018). GWAS with heterogeneous data: estimating the fraction of phenotypic variation mediated by gene expression data. G3 8, 3059-3068.
DOI URL |
| [69] | Schiessl SV, Quezada-Martinez D, Tebartz E, Snowdon RJ, Qian LW (2019). The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci Rep 9, 14911. |
| [70] | Schwechheimer C (2012). Gibberellin signaling in plants- the extended version. Front Plant Sci 2, 107. |
| [71] |
Searle I, He YH, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20, 898-912.
DOI URL |
| [72] |
Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC (2010). JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24, 986-991.
DOI URL |
| [73] |
Sharma N, Geuten K, Giri BS, Varma A (2020). The molecular mechanism of vernalization in Arabidopsis and cereals: role of flowering locus c and its homologs. Physiol Plant 170, 373-383.
DOI URL |
| [74] |
Sharma N, Ruelens P, D'hauw M, Maggen T, Dochy N, Torfs S, Kaufmann K, Rohde A, Geuten K (2017). A flowering locus c homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol 173, 1301-1315.
DOI PMID |
| [75] |
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445-458.
PMID |
| [76] |
Shin JH, Chekanova JA (2014). Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 10, e1004612.
DOI URL |
| [77] |
Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138, 1163-1173.
DOI URL |
| [78] |
Shu K, Chen F, Zhou WG, Luo XF, Dai YJ, Shuai HW, Yang WY (2018). ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol Biol Rep 45, 2727-2731.
DOI URL |
| [79] |
Shu K, Chen Q, Wu YR, Liu RJ, Zhang HW, Wang SF, Tang SY, Yang WY, Xie Q (2016). ABSCISIC ACID- INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J Exp Bot 67, 195-205.
DOI URL |
| [80] |
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777-787.
PMID |
| [81] |
Son GH, Park BS, Song JT, Seo HS (2014). FLC-mediated flowering repression is positively regulated by SUMOylation. J Exp Bot 65, 339-351.
DOI URL |
| [82] |
Srikanth A, Schmid M (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68, 2013-2037.
DOI PMID |
| [83] |
Steffen A, Elgner M, Staiger D (2019). Regulation of flowering time by the RNA-binding proteins AtGRP7 and AtGRP8. Plant Cell Physiol 60, 2040-2050.
DOI URL |
| [84] |
Sung S, Amasino RM (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159-164.
DOI URL |
| [85] |
Sung S, Schmitz RJ, Amasino RM (2006). A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20, 3244-3248.
DOI URL |
| [86] |
Swiezewski S, Crevillen P, Liu FQ, Ecker JR, Jerzmanowski A, Dean C (2007). Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA 104, 3633-3638.
DOI URL |
| [87] |
Swiezewski S, Liu FQ, Magusin A, Dean C (2009). Cold- induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462, 799-802.
DOI URL |
| [88] |
Tadege M, Sheldon CC, Helliwell CA, Stoutjesdijk P, Dennis ES, Peacock WJ (2001). Control of flowering time by FLC orthologues in Brassica napus. Plant J 28, 545-553.
PMID |
| [89] |
Tadege M, Sheldon CC, Helliwell CA, Upadhyaya NM, Dennis ES, Peacock WJ (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol J 1, 361-369.
DOI URL |
| [90] |
Takada S, Akter A, Itabashi E, Nishida N, Shea DJ, Miyaji N, Mehraj H, Osabe K, Shimizu M, Takasaki-Yasuda T, Kakizaki T, Okazaki K, Dennis ES, Fujimoto R (2019). The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci Rep 9, 13843.
DOI URL |
| [91] |
Taylor JL, Massiah A, Kennedy S, Hong YG, Jackson SD (2017). FLC expression is down-regulated by cold treatment in Diplotaxis tenuifolia (wild rocket), but flowering time is unaffected. J Plant Physiol 214, 7-15.
DOI URL |
| [92] | Teng YB, Liang Y, Wang MY, Mai HC, Ke LP (2019). Nitrate Transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS C in Arabidopsis thaliana. Plant Sci 284, 30-36. |
| [93] | Tian YK, Zheng H, Zhang F, Wang SL, Ji XR, Xu C, He YH, Ding Y (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci Adv 5, eaau7246. |
| [94] | Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3, e86. |
| [95] | Wang HLV, Chekanova JA (2017). Long noncoding RNAs in plants. Adv Exp Med Biol 1008, 133-154. |
| [96] |
Wang JW (2014). Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65, 4723-4730.
DOI URL |
| [97] |
Wang YP, Li L, Ye TT, Lu YM, Chen X, Wu Y (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64, 675-684.
DOI URL |
| [98] |
Wang YZ, Gu XF, Yuan WY, Schmitz RJ, He YH (2014). Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 28, 727-736.
DOI URL |
| [99] |
Whittaker C, Dean C (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol 33, 555-575.
DOI PMID |
| [100] |
Wilkinson KA, Henley JM (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428, 133-145.
DOI PMID |
| [101] |
Wu Z, Fang XF, Zhu DL, Dean C (2020). Autonomous pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol 182, 27-37.
DOI URL |
| [102] | Xiao J, Zhang H, Xing LJ, Xu SJ, Liu HH, Chong K, Xu YY (2013). Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 170, 444-451. |
| [103] |
Xiong F, Ren JJ, Yu Q, Wang YY, Lu CC, Kong LJ, Otegui MS, Wang XL (2019). AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol 223, 277-292.
DOI PMID |
| [104] | Xu ML, Hu TQ, Zhao JF, Park MY, Earley KW, Wu G, Yang L, Poethig RS (2016). Developmental functions of miR156- regulated SQUAMOSA PROMOTER BINDING PROTEIN- LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet 12, e1006263. |
| [105] |
Yan FH, Zhang LP, Cheng F, Yu DM, Hu JY (2021). Accession-specific flowering time variation in response to nitrate fluctuation in Arabidopsis thaliana. Plant Divers 43, 78-85.
DOI URL |
| [106] |
Yan YY, Shen LS, Chen Y, Bao SJ, Thong ZH, Yu H (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev Cell 30, 437-448.
DOI URL |
| [107] |
Yang HC, Han ZF, Cao Y, Fan D, Li H, Mo HX, Feng Y, Liu L, Wang Z, Yue YL, Cui SJ, Chen S, Chai JJ, Ma LG (2012). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet 8, e1002664.
DOI URL |
| [108] |
Yang HC, Howard M, Dean C (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24, 1793-1797.
DOI URL |
| [109] |
Zhang L, Jiménez-Gómez JM (2020). Functional analysis of FRIGIDA using naturally occurring variation in Arabidopsis thaliana. Plant J 103, 154-165.
DOI URL |
| [110] |
Zhao T, Ni ZF, Dai Y, Yao YY, Nie XL, Sun QX (2006). Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.). Mol Genet Genomics 276, 334-350.
PMID |
| [111] |
Zhao YS, Zhu P, Hepworth J, Bloomer R, Antoniou- Kourounioti RL, Doughty J, Heckmann A, Xu CY, Yang HC, Dean C (2021). Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev 35, 888-898.
DOI URL |
| [112] |
Zhao Z, Yu Y, Meyer D, Wu CJ, Shen WH (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7, 1256-1260.
DOI URL |
| [113] |
Zhou JX, Liu ZW, Li YQ, Li L, Wang BJ, Chen S, He XJ (2018). Arabidopsis PWWP domain proteins mediate H3K27 trimethylation on FLC and regulate flowering time. J Integr Plant Biol 60, 362-368.
DOI URL |
| [114] |
Zhu DL, Rosa S, Dean C (2015). Nuclear organization changes and the epigenetic silencing of FLC during vernalization. J Mol Biol 427, 659-669.
DOI URL |
| [1] | 杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础[J]. 植物学报, 2024, 59(3): 373-382. |
| [2] | 杨小凤, 李小蒙, 廖万金. 植物开花时间的遗传调控通路研究进展[J]. 生物多样性, 2021, 29(6): 825-842. |
| [3] | 李琳, 路宁娜, 樊宝丽, 赵志刚. 雌雄异熟植物露蕊乌头开花时间对雌雄功能期及表型性别的影响[J]. 生物多样性, 2016, 24(6): 665-671. |
| [4] | 张艺能, 周玉萍, 陈琼华, 黄小玲, 田长恩. 拟南芥开花时间调控的分子基础[J]. 植物学报, 2014, 49(4): 469-482. |
| [5] | 刘乐乐, 刘左军, 杜国祯, 赵志刚. 毛茛状金莲花不同花期的花特征和访花昆虫的变化及表型选择[J]. 生物多样性, 2012, 20(3): 317-323. |
| [6] | 罗睿*;郭建军. 植物开花时间: 自然变异与遗传分化[J]. 植物学报, 2010, 45(01): 109-118. |
| [7] | 洪薇 曹家树. FLC基因表达在植物春化过程中的作用[J]. 植物学报, 2002, 19(04): 406-411. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||