

植物学报 ›› 2018, Vol. 53 ›› Issue (5): 625-633.DOI: 10.11983/CBB17169 cstr: 32102.14.CBB17169
收稿日期:2017-09-06
接受日期:2017-11-16
出版日期:2018-09-01
发布日期:2018-11-29
通讯作者:
白洁
作者简介:
作者简介: 路安民(图中左), 植物系统分类学家。20世纪60-70年代编著《中国植物志》等, 后从事植物系统发育和进化研究。“七五”以来主持了4项中科院、国家自然科学基金委重大和重点项目。1991年获国务院颁发的有突出贡献科学家荣誉证书。1987年8月-1990年12月担任中科院植物所所长。
基金资助:
Liu Wei1, Tong Yong’ao1,2, Bai Jie1,*( )
)
Received:2017-09-06
Accepted:2017-11-16
Online:2018-09-01
Published:2018-11-29
Contact:
Bai Jie
About author:† These authors contributed equally to this paper
摘要: tRNA片段(tRF)是tRNA通过非随机剪切产生的RNA片段, 其产生和功能机制尚不明确; 而在水稻(Oryza sativa)雄配子体发育过程中, 人们对tRNA更是知之甚少。通过高通量测序, 在水稻雄配子体发育过程中发现了长度范围较大的tRFs; 进一步采用logo对tRFs两端的序列进行分析, 发现了4个有序列特征(其中3个未见报道)和1个无序列特征的酶切位点; 通过NCBI Blast预测了tRF靶基因, 发现其大多靶向转座因子。研究结果对揭示tRF产生机制以及水稻雄配子体发育研究有一定的参考价值。
刘魏, 童永鳌, 白洁. 水稻雄配子体发育过程中tRNA片段的生物信息学分析. 植物学报, 2018, 53(5): 625-633.
Liu Wei, Tong Yong’ao, Bai Jie. Bioinformatics Analysis of tRNA-derived Fragments in Rice Male Gametophyte Development. Chinese Bulletin of Botany, 2018, 53(5): 625-633.
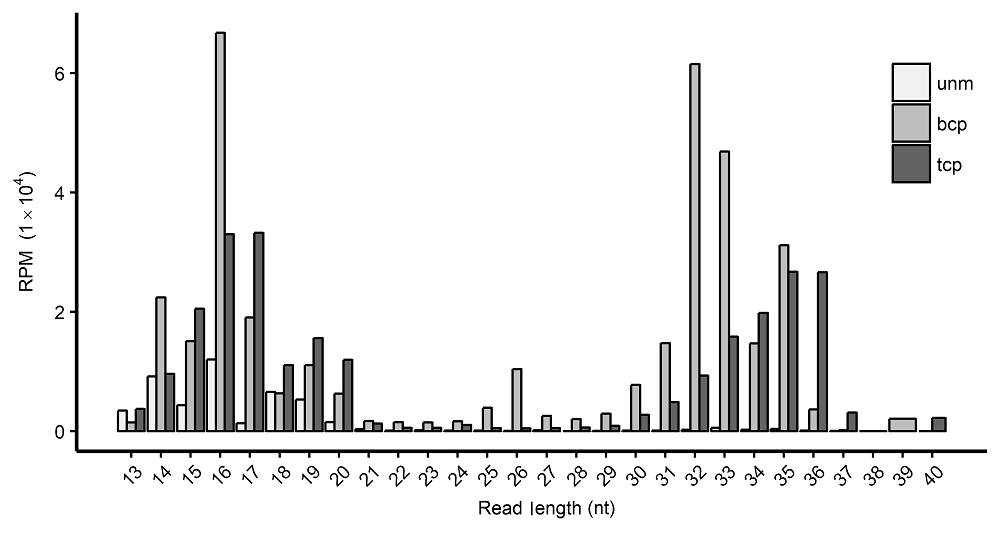
图1 水稻雄配子三类花粉中的tRF长度分布 unm: 单核小孢子; bcp: 二细胞花粉; tcp: 三细胞花粉; RPM: 每百万条读段里的(目标)读段数
Figure 1 Length distribution of tRFs in three types of pollens in rice male gameteunm: Uni-nucleate microspore; bcp: Bi-cellular pollen; tcp : Tri-cellular pollen; RPM: Reads per million
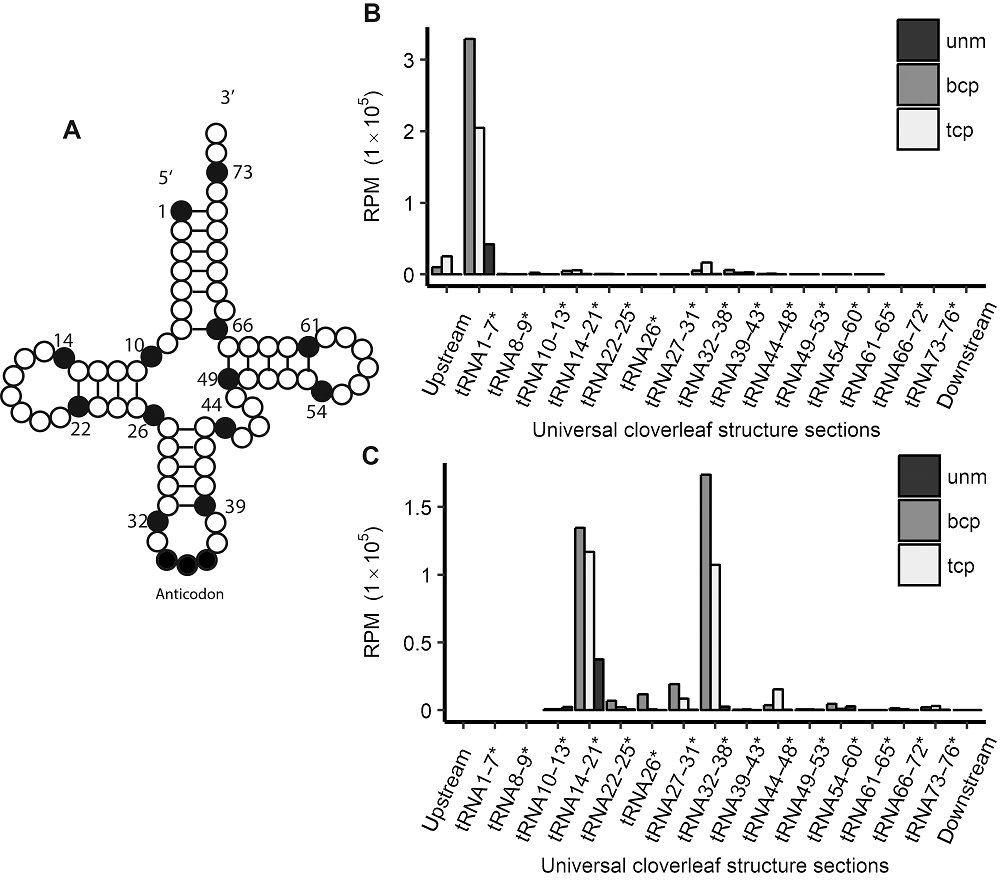
图2 水稻雄配子三类花粉的tRF起始和终止位点在四叶草结构中的分布 (A) tRNA二级结构示意图; (B) tRF起点分布; (C) tRF终点分布。*代表图2A中数字的位置, 不代表碱基编号。unm、bcp、tcp及RPM同图1。
Figure 2 tRFs’ starting and ending site distribution in tRNAs’ universal cloverleaf structure sections in three types of pollens in rice male gamete(A) Schematic view of tRNAs’ cloverleaf-like secondary structure; (B) tRFs’ starting site positions in tRNA; (C) tRFs’ ending site positions in tRNA. * indicate that the numbers before can only be regarded as a positions represented in Figure 2A instead of base ID. unm, bcp, tcp and RPM see Figure 1.
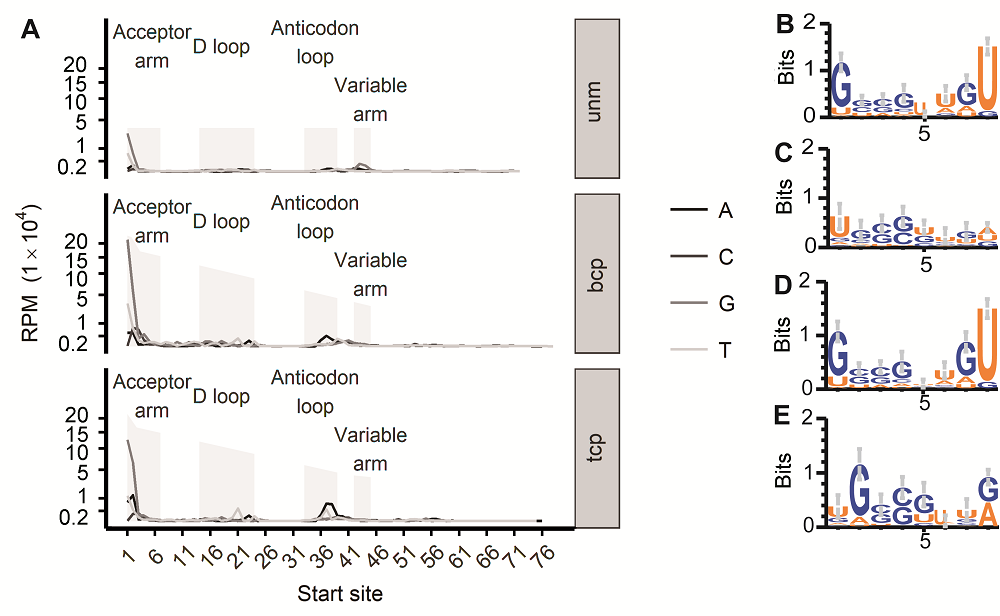
图3 水稻雄配子三类花粉的tRF 5’端碱基分布偏好性(A) 起始于tRNA各个位置的tRF起点碱基分布情况(纵坐标各点相对位置进行了1/2次幂变形处理); (B) bcp样品中tRF起始于tRNA第1位时, 对应宿主tRNA前8个碱基logo图; (C) bcp样品中tRF起始于tRNA第2位时, 对应宿主tRNA前8个碱基logo图; (D) tcp样品中tRF起始于tRNA第1位时, 对应宿主tRNA前8个碱基logo图; (E) tcp样品中tRF起始于tRNA第2位时, 对应宿主tRNA前8个碱基logo图。unm、bcp、tcp和RPM同图1。
Figure 3 tRFs’ 5’ end base bias in three types of pollens in rice male gamete(A) tRF starting site’s distribution in each position of their host tRNA (The vertical axis labels’ positions were transferred according to their square root number); (B) First 8 base logo of the tRNAs that produce tRFs from the 1st base in bcp; (C) First 8 base logo of tRNAs that produce tRFs from the 2nd base in bcp; (D) First 8 base logo of the tRNAs that produce tRFs from the 1st base in tcp; (E) First 8 base logo of tRNAs that produce tRFs from the 2nd base in tcp. unm, bcp, tcp and RPM see Figure 1.
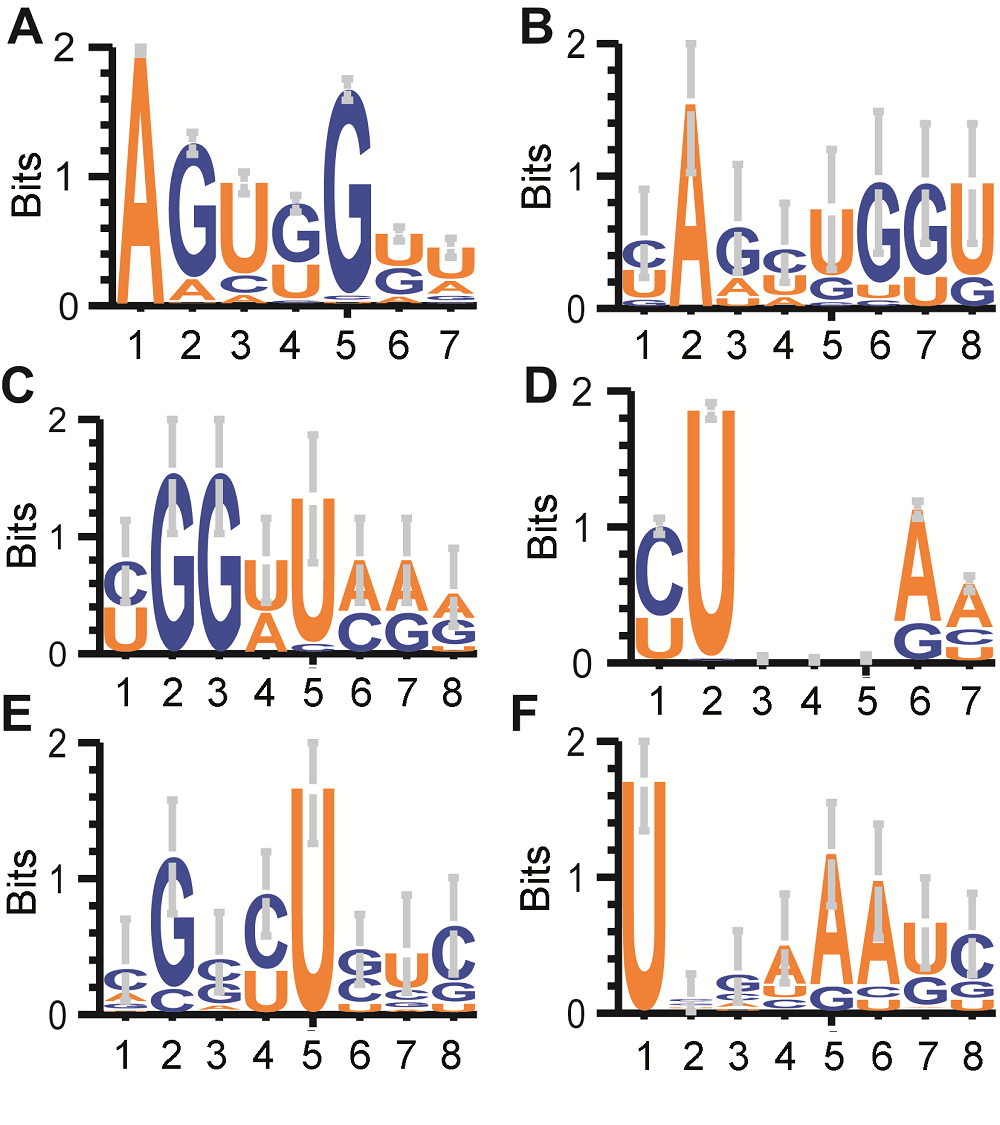
图4 水稻雄配子三类花粉的tRNA剪切位置周围碱基分布(A), (D) 所有tRNA的D环和反密码子环上前7个碱基分布logo图; (B) bcp于D环第3个碱基终止的tRF对应tRNA序列logo图; (C) tcp于D环第7个碱基起始的tRF对应tRNA序列logo图; (E) bcp于反密码子环第1个碱基终止的tRF对应tRNA序列logo图; (F) tcp于反密码子环第6个碱基起始的tRF对应tRNA序列logo图。除了图A和D, 各图横轴4和5之间为“剪切发生位置”。
Figure 4 Sequence motif logo for tRNA 8 mer around cleavage sites in three types of pollens in rice male gamete(A), (D) Sequence logo of all tRNA’s first 7 bases of D loop and Anticodon loop; (B) For bcp tRFs ended at the 3rd base of D loop; (C) For tcp tRFs started at the 7th base of D loop; (E) For bcp tRFs ended at the 1st base of Anticodon loop; (F) For tcp tRFs started at the 6th base of Anticodon loop. The position between 4 and 5 in X axis of Figure B, C, E and F stand for cleavage site.
| Sample | No. of non-TE genes | No. of genic TEs | No. of intergenic TEs | TE Percentage (%) |
|---|---|---|---|---|
| unm | 317 | 166 | 506 | 67.95 |
| bcp | 728 | 403 | 905 | 64.24 |
| tcp | 529 | 287 | 736 | 65.91 |
表1 被靶向的非TE基因和TE数的对比
Table 1 tRFs targeted loci of non-TE and TE genes
| Sample | No. of non-TE genes | No. of genic TEs | No. of intergenic TEs | TE Percentage (%) |
|---|---|---|---|---|
| unm | 317 | 166 | 506 | 67.95 |
| bcp | 728 | 403 | 905 | 64.24 |
| tcp | 529 | 287 | 736 | 65.91 |
| 1 | Abe T, Ikemura T, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, Inokuchi H (2009). tRNADB-CE: tRNA gene database curated manually by experts.Nucleic Acids Res 37, D163-D168. |
| 2 | Alves CS, Vicentini R, Duarte GT, Pinoti VF, Vincentz M, Nogueira FTS (2016). Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants.Plant Mol Biol 93, 35-48. |
| 3 | Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou XP, Russell SD, Sundaresan V (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76, 729-741. |
| 4 | Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D (2011). Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus.RNA Biol 8, 538-547. |
| 5 | Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G (2009). Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs.RNA 15, 2147-2160. |
| 6 | Couvillion MT, Sachidanandam R, Collins K (2010). A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24, 2742-2747. |
| 7 | Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM (2006). MODOMICS: a database of RNA modification pathways.Nucleic Acids Res 34, D145-D149. |
| 8 | Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P (2010). Angiogenin- induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly.J Biol Chem 285, 10959-10968. |
| 9 | Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N (2017). A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress.RNA Biol 14, 1364-1373. |
| 10 | Goodarzi H, Liu XH, Nguyen HCB, Zhang S, Fish L, Tavazoie SF (2015). Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement.Cell 161, 790-802. |
| 11 | Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H (2009). MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10, 643. |
| 12 | Hsieh LC, Lin SI, Kuo HF, Chiou TJ (2010). Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots.Plant Signal Behav 5, 537-539. |
| 13 | Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A (2015). Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct 10, 51. |
| 14 | Keam SP, Sobala A, Ten Have S, Hutvagner G (2017). tRNA-Derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation.J Proteome Res 16, 413-420. |
| 15 | Kumar P, Anaya J, Mudunuri SB, Dutta A (2014). Meta- analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets.BMC Biol 12, 78. |
| 16 | Kumar P, Kuscu C, Dutta A (2016). Biogenesis and function of transfer RNA-related fragments (tRFs).Trends Biochem Sci 41, 679-689. |
| 17 | Loss-Morais G, Waterhouse PM, Margis R (2013). Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets.Biol Direct 8, 6. |
| 18 | Lowe TM, Eddy SR (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence.Nucleic Acids Res 25, 955-964. |
| 19 | Martinez G, Choudury SG, Slotkin RK (2017). tRNA- derived small RNAs target transposable element transcripts.Nucleic Acids Res 45, 5142-5152. |
| 20 | Nawrot B, Malkiewicz A, Smith WS, Sierzputowska-Gracz H, Agris PF (1995). RNA modified uridines VII: chemical synthesis and initial analysis of tRNA D-Loop oligomers with tandem modified uridines.Nucleos Nucleot Nud 14, 143-165. |
| 21 | Peng H, Chun J, Ai TB, Tong YA, Zhang R, Zhao MM, Chen F, Wang SH (2012). MicroRNA profiles and their control of male gametophyte development in rice.Plant Mol Biol 80, 85-102. |
| 22 | Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage.Genes Dev 24, 1590-1595. |
| 23 | Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun FY, Song LN, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals.Science 351, 391-396. |
| 24 | Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijo JA, Martienssen RA (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen.Cell 136, 461-472. |
| 25 | Sobala A, Hutvagner G (2013). Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells.RNA Biol 10, 553-563. |
| 26 | Stroud H, Ding B, Simon SA, Feng SH, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE (2013). Plants regenerated from tissue culture contain stable epigenome changes in rice.eLife 2, e00354. |
| 27 | Thompson DM, Parker R (2009). The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185, 43-50. |
| 28 | Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis.Nat Struct Mol Biol 19, 900-905. |
| 29 | Vare VYP, Eruysal ER, Narendran A, Sarachan KL, Agris PF (2017). Chemical and conformational diversity of modified nucleosides affects tRNA structure and function.Bio- molecules 7, 29. |
| 30 | Wang F, Johnson NR, Coruh C, Axtell MJ (2016a). Genome-wide analysis of single non-templated nucleotides in plant endogenous siRNAs and miRNAs.Nucleic Acids Res 44, 7395-7405. |
| 31 | Wang QH, Li TT, Xu K, Zhang W, Wang XL, Quan JL, Jin WB, Zhang MX, Fan GJ, Wang MB, Shan WX (2016b). The tRNA-derived small RNAs regulate gene expression through triggering sequence-specific degradation of target transcripts in the oomycete pathogen Phytophthora sojae. Front Plant Sci 7, 1938. |
| 32 | Wei LQ, Xu WY, Deng ZY, Su Z, Xue YB, Wang T (2010). Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11, 338. |
| 33 | Wei LQ, Yan LF, Wang T (2011). Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol 12, R53. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析[J]. 植物学报, 2025, 60(3): 377-392. |
| [4] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [5] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [6] | 巴苏艳, 赵春艳, 刘媛, 方强. 通过虫体花粉识别构建植物‒传粉者网络: 人工模型与AI模型高度一致[J]. 生物多样性, 2024, 32(6): 24088-. |
| [7] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [8] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [9] | 覃思颖, 罗燕, 张禾, 胡君, 廖菊够. 花粉管细胞壁原子力显微镜观测制样方法优化[J]. 植物学报, 2024, 59(5): 783-791. |
| [10] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [11] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [12] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [13] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [14] | 吕晓琴, 李杨, 王顺雨, 姚仁秀, 王晓月. 中甸乌头总状花序不同位置花粉和花蜜的化学性状没有显著差异[J]. 生物多样性, 2024, 32(1): 23371-. |
| [15] | 张飞飞, 杨天凤, 陈莉荣, 刘冬梅, 杨柳园, 杨杜宇, 鞠鹏, 陆露. 被子植物花粉颜色多样性及应用研究进展[J]. 生物多样性, 2024, 32(1): 23346-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||