

植物学报 ›› 2018, Vol. 53 ›› Issue (5): 612-624.DOI: 10.11983/CBB17117 cstr: 32102.14.CBB17117
赵雪惠1,2, 王庆杰1,2, 李晨1,2, 陈修德1,2, 肖伟1,2, 高东升1,2, 付喜玲1,2,*( )
)
收稿日期:2017-06-15
接受日期:2017-10-16
出版日期:2018-09-01
发布日期:2018-11-29
通讯作者:
付喜玲
作者简介:
作者简介: 路安民(图中左), 植物系统分类学家。20世纪60-70年代编著《中国植物志》等, 后从事植物系统发育和进化研究。“七五”以来主持了4项中科院、国家自然科学基金委重大和重点项目。1991年获国务院颁发的有突出贡献科学家荣誉证书。1987年8月-1990年12月担任中科院植物所所长。
基金资助:
Zhao Xuehui1,2, Wang Qingjie1,2, Li Chen1,2, Chen Xiude1,2, Xiao Wei1,2, Gao Dongsheng1,2, Fu Xiling1,2,*( )
)
Received:2017-06-15
Accepted:2017-10-16
Online:2018-09-01
Published:2018-11-29
Contact:
Fu Xiling
About author:† These authors contributed equally to this paper
摘要: 以中油四号油桃(Prunus persica var. nectarina)为研究对象, 利用MEGA 6.0、MEME、GSDS和DNAMAN 6.0等软件对桃ERF家族数据进行生物信息学分析, 鉴定得到102个ERF转录因子家族基因, 并通过构建系统进化树将这102个基因分为10个子家族(I-X)。基因结构分析表明, 有81个基因不含内含子, 20个基因含有1个内含子, 有1个基因与其它成员差异较大, 含有5个内含子。保守元件分析表明, ERF家族包含20个保守元件, 其中Motif 1、Motif 2和Motif 4都属于AP2/ERF结构域, 同一个保守元件主要出现在同一个子家族中, 并且大部分保守元件的功能未知。VIII子家族基因的荧光定量PCR分析表明, 在桃叶芽处于不同的发育状态时, PpeERF068的表达量存在较大差异, 光照培养箱中培养的桃芽在萌发过程中各时期表达量变化趋势进一步表明该基因可能与叶芽萌发有关, 将其命名为PpeEBB1。该研究为进一步揭示PpeEBB1的分子机制奠定了基础, 并为桃树的栽培管理和熟期调控了提供理论指导。
赵雪惠, 王庆杰, 李晨, 陈修德, 肖伟, 高东升, 付喜玲. 桃ERF转录因子家族生物信息学分析及 芽萌发相关基因筛选. 植物学报, 2018, 53(5): 612-624.
Zhao Xuehui, Wang Qingjie, Li Chen, Chen Xiude, Xiao Wei, Gao Dongsheng, Fu Xiling. Genome-wide Identification of Ethylene Responsive Factor (ERF) Family Genes in Peach and Screening of Genes Related to Germination. Chinese Bulletin of Botany, 2018, 53(5): 612-624.
| Locus name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Prupe.3g094700 | GCGGAGATAAGAGACCGAAT | AGAAGATGATGAGCAAGGTGAG |
| Prupe.1g245500 | AGTATCACAGACGACGCAGG | ATCGCCCATCACTCAAGA |
| Prupe.1g212700 | TATGCGGCGGAGATAAGA | GGGTGAGATAATGGAGGTGA |
| Prupe.1g139600 | TGGATTCTGGGACAACCA | GGAACTGATGGAGAGCAAGAG |
| Prupe.4g176200 | GCCCAAACTCCAAAGAGA | GTTGTTGTGGCTGTTGATGT |
| Prupe.4g051400 | TCTCGCTCAGGGTGTTAGTA | CAGCCGTTGATGAAACTG |
| Prupe.8g230600 | TTGCCTCACACGCTTGTTA | GAACGGTTTCTTCTCTTTCCTC |
| Prupe.4g051200 | TCTCTCTCTGGACCTCAACAT | CGCCAACATTTCCTACAAC |
| Prupe.4g222300 | AAGCCAAGACCAACTTCCC | TGTCCTGAAACCCGTAACC |
| Prupe.3g209100 | TCCTCATCCTCCGTCGTAGA | ATCACCACAAACCTCACCG |
| Actin | GTTATTCTTCATCGGCGTCTTCG | CTTCACCATTCCAGTTCCATTGTC |
表1 实时荧光定量PCR引物序列
Table 1 Primers of qRT-PCR
| Locus name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Prupe.3g094700 | GCGGAGATAAGAGACCGAAT | AGAAGATGATGAGCAAGGTGAG |
| Prupe.1g245500 | AGTATCACAGACGACGCAGG | ATCGCCCATCACTCAAGA |
| Prupe.1g212700 | TATGCGGCGGAGATAAGA | GGGTGAGATAATGGAGGTGA |
| Prupe.1g139600 | TGGATTCTGGGACAACCA | GGAACTGATGGAGAGCAAGAG |
| Prupe.4g176200 | GCCCAAACTCCAAAGAGA | GTTGTTGTGGCTGTTGATGT |
| Prupe.4g051400 | TCTCGCTCAGGGTGTTAGTA | CAGCCGTTGATGAAACTG |
| Prupe.8g230600 | TTGCCTCACACGCTTGTTA | GAACGGTTTCTTCTCTTTCCTC |
| Prupe.4g051200 | TCTCTCTCTGGACCTCAACAT | CGCCAACATTTCCTACAAC |
| Prupe.4g222300 | AAGCCAAGACCAACTTCCC | TGTCCTGAAACCCGTAACC |
| Prupe.3g209100 | TCCTCATCCTCCGTCGTAGA | ATCACCACAAACCTCACCG |
| Actin | GTTATTCTTCATCGGCGTCTTCG | CTTCACCATTCCAGTTCCATTGTC |
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
|---|---|---|---|---|---|---|---|
| I | PpeERF#001 | Prupe.6g182200 | 1122 | 373 | 41331.3 | 6.56 | 13 |
| I | PpeERF#002 | Prupe.3g009400 | 642 | 213 | 23101.2 | 9.79 | 7 |
| I | PpeERF#003 | Prupe.1g513600 | 1137 | 378 | 41236.8 | 6.37 | 9 |
| I | PpeERF#004 | Prupe.5g117800 | 933 | 310 | 34377.6 | 8.34 | 9 |
| I | PpeERF#005 | Prupe.3g157100 | 1386 | 461 | 51713.0 | 5.97 | 0 |
| I | PpeERF#006 | Prupe.1g008400 | 1230 | 409 | 45352.9 | 7.81 | 2 |
| I | PpeERF#007 | Prupe.8g198700 | 696 | 231 | 26301.6 | 9.19 | 0 |
| II | PpeERF#008 | Prupe.6g176400 | 540 | 179 | 20308.7 | 9.51 | 0 |
| II | PpeERF#009 | Prupe.1g432000 | 528 | 175 | 19844.9 | 9.39 | 1 |
| II | PpeERF#010 | Prupe.1g464400 | 453 | 150 | 17240.2 | 9.42 | 2 |
| II | PpeERF#011 | Prupe.6g231500 | 501 | 166 | 17849.1 | 8.72 | 0 |
| II | PpeERF#012 | Prupe.1g546100 | 501 | 166 | 17890.7 | 5.62 | 0 |
| II | PpeERF#013 | Prupe.1g500900 | 726 | 241 | 26308.4 | 5.61 | 0 |
| II | PpeERF#014 | Prupe.7g194400 | 648 | 215 | 23980.2 | 5.16 | 0 |
| II | PpeERF#015 | Prupe.1g001700 | 552 | 183 | 20044.5 | 7.74 | 0 |
| III | PpeERF#016 | Prupe.5g090700 | 558 | 185 | 19627.0 | 4.89 | 0 |
| III | PpeERF#017 | Prupe.5g090600 | 537 | 178 | 19407.7 | 5.67 | 0 |
| III | PpeERF#018 | Prupe.5g090400 | 543 | 180 | 19547.0 | 5.50 | 0 |
| III | PpeERF#019 | Prupe.5g090300 | 543 | 180 | 19510.8 | 5.50 | 0 |
| III | PpeERF#020 | Prupe.5g090800 | 555 | 184 | 19705.9 | 4.64 | 0 |
| III | PpeERF#021 | Prupe.2g289600 | 696 | 231 | 24485.1 | 4.83 | 0 |
| III | PpeERF#022 | Prupe.7g116300 | 609 | 202 | 21536.7 | 4.91 | 0 |
| III | PpeERF#023 | Prupe.2g190200 | 609 | 202 | 22713.9 | 5.87 | 1 |
| III | PpeERF#024 | Prupe.2g123200 | 831 | 276 | 30098.8 | 4.94 | 0 |
| III | PpeERF#025 | Prupe.5g065300 | 762 | 253 | 28183.5 | 4.91 | 0 |
| III | PpeERF#026 | Prupe.1g448600 | 771 | 256 | 27960.9 | 5.11 | 0 |
| III | PpeERF#027 | Prupe.7g222700 | 789 | 262 | 28536.4 | 4.94 | 0 |
| III | PpeERF#028 | Prupe.3g062800 | 732 | 243 | 26436.2 | 6.67 | 1 |
| III | PpeERF#029 | Prupe.7g115900 | 606 | 201 | 22481.2 | 4.95 | 0 |
| III | PpeERF#030 | Prupe.2g289500 | 690 | 229 | 24976.9 | 5.14 | 7 |
| III | PpeERF#031 | Prupe.5g090500 | 723 | 240 | 27217.8 | 7.69 | 0 |
| III | PpeERF#032 | Prupe.5g090100 | 711 | 236 | 26352.7 | 5.17 | 0 |
| III | PpeERF#033 | Prupe.5g090000 | 693 | 230 | 25658.8 | 5.13 | 7 |
| III | PpeERF#034 | Prupe.5g090200 | 699 | 232 | 26157.5 | 5.03 | 1 |
| III | PpeERF#035 | Prupe.5g089900 | 693 | 230 | 25521.9 | 7.78 | 0 |
| III | PpeERF#036 | Prupe.1g545700 | 522 | 173 | 19078.3 | 5.44 | 0 |
| III | PpeERF#037 | Prupe.1g545400 | 600 | 199 | 22101.0 | 5.54 | 0 |
| IV | PpeERF#038 | Prupe.2g253000 | 1041 | 346 | 37474.1 | 6.52 | 0 |
| IV | PpeERF#039 | Prupe.6g284400 | 1554 | 517 | 58370.7 | 4.74 | 0 |
| IV | PpeERF#040 | Prupe.2g256900 | 1167 | 388 | 42473.1 | 4.64 | 3 |
| IV | PpeERF#041 | Prupe.3g223300 | 591 | 196 | 21305.5 | 8.32 | 0 |
| IV | PpeERF#042 | Prupe.7g066700 | 858 | 285 | 32054.9 | 5.73 | 0 |
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
| IV | PpeERF#043 | Prupe.6g354000 | 672 | 223 | 23337.9 | 8.50 | 0 |
| IV | PpeERF#044 | Prupe.1g372100 | 768 | 255 | 28137.6 | 6.55 | 0 |
| V | PpeERF#045 | Prupe.3g096000 | 639 | 212 | 24310.2 | 5.94 | 0 |
| V | PpeERF#046 | Prupe.1g390800 | 588 | 195 | 22413.4 | 6.66 | 6 |
| V | PpeERF#047 | Prupe.1g480400 | 672 | 223 | 24759.7 | 9.10 | 0 |
| V | PpeERF#048 | Prupe.7g243600 | 732 | 243 | 26570.6 | 7.02 | 0 |
| V | PpeERF#049 | Prupe.3g084600 | 567 | 188 | 20855.3 | 5.97 | 0 |
| V | PpeERF#050 | Prupe.7g004900 | 945 | 314 | 34636.3 | 5.44 | 0 |
| V | PpeERF#051 | Prupe.6g004400 | 867 | 288 | 31688.5 | 6.01 | 0 |
| V | PpeERF#052 | Prupe.3g263000 | 1275 | 424 | 46904.3 | 4.58 | 0 |
| V | PpeERF#053 | Prupe.5g136200 | 939 | 312 | 33152.5 | 6.13 | 0 |
| V | PpeERF#054 | Prupe.1g084800 | 936 | 311 | 33626.0 | 8.92 | 0 |
| V | PpeERF#055 | Prupe.5g213800 | 675 | 224 | 25367.4 | 8.71 | 0 |
| VI | PpeERF#056 | Prupe.5g114100 | 1002 | 333 | 37019.0 | 4.98 | 0 |
| VI | PpeERF#057 | Prupe.2g306400 | 1098 | 365 | 41098.9 | 4.75 | 2 |
| VI | PpeERF#058 | Prupe.2g183200 | 864 | 287 | 32269.0 | 4.95 | 0 |
| VI | PpeERF#059 | Prupe.3g019900 | 570 | 189 | 20614.4 | 9.37 | 0 |
| VI | PpeERF#060 | Prupe.7g060700 | 588 | 195 | 21933.2 | 10.37 | 2 |
| VI | PpeERF#061 | Prupe.6g039700 | 927 | 308 | 34031.9 | 5.49 | 2 |
| VI | PpeERF#062 | Prupe.5g220700 | 1311 | 436 | 49386.8 | 6.25 | 1 |
| VI | PpeERF#063 | Prupe.1g310100 | 1020 | 339 | 38021.2 | 4.92 | 1 |
| VII | PpeERF#064 | Prupe.8g264900 | 966 | 321 | 35682.6 | 5.68 | 4 |
| VII | PpeERF#065 | Prupe.3g032300 | 1149 | 382 | 42495.9 | 4.81 | 84 |
| VII | PpeERF#066 | Prupe.1g130300 | 870 | 289 | 31564.0 | 7.66 | 31 |
| VIII | PpeERF#067 | Prupe.3g094700 | 870 | 289 | 31781.0 | 9.56 | 0 |
| VIII | PpeERF#068 | Prupe.1g245500 | 1275 | 424 | 46297.0 | 7.73 | 0 |
| VIII | PpeERF#069 | Prupe.1g212700 | 831 | 276 | 29938.4 | 4.94 | 0 |
| VIII | PpeERF#070 | Prupe.1g139600 | 1092 | 363 | 39938.7 | 5.60 | 0 |
| VIII | PpeERF#071 | Prupe.4g176200 | 687 | 228 | 24245.0 | 9.62 | 2 |
| VIII | PpeERF#072 | Prupe.4g051400 | 795 | 264 | 28447.6 | 9.47 | 1 |
| VIII | PpeERF#073 | Prupe.8g230600 | 546 | 181 | 19951.0 | 9.99 | 0 |
| VIII | PpeERF#074 | Prupe.4g051200 | 519 | 172 | 18303.7 | 9.20 | 3 |
| VIII | PpeERF#075 | Prupe.4g222300 | 696 | 231 | 25022.8 | 7.62 | 20 |
| VIII | PpeERF#076 | Prupe.3g209100 | 708 | 235 | 25471.3 | 6.58 | 3 |
| IX | PpeERF#077 | Prupe.2g129700 | 672 | 223 | 24950.8 | 5.41 | 0 |
| IX | PpeERF#078 | Prupe.2g129500 | 651 | 216 | 23968.9 | 5.99 | 0 |
| IX | PpeERF#079 | Prupe.2g129600 | 714 | 237 | 25948.7 | 5.47 | 0 |
| IX | PpeERF#080 | Prupe.2g129300 | 816 | 271 | 29817.0 | 5.17 | 0 |
| IX | PpeERF#081 | Prupe.2g129400 | 621 | 206 | 22559.1 | 5.97 | 0 |
| IX | PpeERF#082 | Prupe.5g061800 | 846 | 281 | 30157.0 | 9.09 | 10 |
| IX | PpeERF#083 | Prupe.6g064700 | 2115 | 704 | 77761.3 | 9.77 | 15 |
| IX | PpeERF#084 | Prupe.2g272300 | 783 | 260 | 29210.8 | 6.53 | 3 |
| IX | PpeERF#085 | Prupe.5g062000 | 1059 | 352 | 39322.2 | 5.90 | 5 |
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
| IX | PpeERF#086 | Prupe.2g272400 | 945 | 314 | 35018.8 | 6.17 | 21 |
| IX | PpeERF#087 | Prupe.2g272500 | 726 | 241 | 27069.2 | 5.92 | 0 |
| IX | PpeERF#088 | Prupe.8g224700 | 540 | 179 | 20470.7 | 6.08 | 0 |
| IX | PpeERF#089 | Prupe.1g037800 | 492 | 163 | 18008.7 | 6.85 | 0 |
| IX | PpeERF#090 | Prupe.8g224800 | 465 | 154 | 17059.7 | 7.79 | 0 |
| IX | PpeERF#091 | Prupe.1g037700 | 417 | 138 | 15204.7 | 6.84 | 0 |
| IX | PpeERF#092 | Prupe.4g055600 | 795 | 264 | 29431.3 | 5.61 | 0 |
| IX | PpeERF#093 | Prupe.4g055500 | 564 | 187 | 20369.6 | 9.51 | 2 |
| IX | PpeERF#094 | Prupe.6g348700 | 744 | 247 | 28089.7 | 4.92 | 0 |
| IX | PpeERF#095 | Prupe.8g224600 | 759 | 252 | 28672.0 | 4.99 | 1 |
| IX | PpeERF#096 | Prupe.1g037900 | 699 | 232 | 25950.8 | 5.54 | 1 |
| X | PpeERF#097 | Prupe.8g125100 | 816 | 271 | 30318.2 | 8.16 | 0 |
| X | PpeERF#098 | Prupe.7g134100 | 801 | 266 | 29255.4 | 7.67 | 0 |
| X | PpeERF#099 | Prupe.5g141200 | 696 | 231 | 25763.5 | 9.21 | 0 |
| X | PpeERF#100 | Prupe.6g165700 | 1215 | 404 | 43314.9 | 7.07 | 2 |
| X | PpeERF#101 | Prupe.5g141300 | 816 | 271 | 30005.0 | 6.16 | 0 |
| X | PpeERF#102 | Prupe.1g214900 | 741 | 246 | 27203.7 | 6.23 | 1 |
表2 桃ERF家族基因及其相关信息
Table 2 List of PpeERF family genes and their related information
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
|---|---|---|---|---|---|---|---|
| I | PpeERF#001 | Prupe.6g182200 | 1122 | 373 | 41331.3 | 6.56 | 13 |
| I | PpeERF#002 | Prupe.3g009400 | 642 | 213 | 23101.2 | 9.79 | 7 |
| I | PpeERF#003 | Prupe.1g513600 | 1137 | 378 | 41236.8 | 6.37 | 9 |
| I | PpeERF#004 | Prupe.5g117800 | 933 | 310 | 34377.6 | 8.34 | 9 |
| I | PpeERF#005 | Prupe.3g157100 | 1386 | 461 | 51713.0 | 5.97 | 0 |
| I | PpeERF#006 | Prupe.1g008400 | 1230 | 409 | 45352.9 | 7.81 | 2 |
| I | PpeERF#007 | Prupe.8g198700 | 696 | 231 | 26301.6 | 9.19 | 0 |
| II | PpeERF#008 | Prupe.6g176400 | 540 | 179 | 20308.7 | 9.51 | 0 |
| II | PpeERF#009 | Prupe.1g432000 | 528 | 175 | 19844.9 | 9.39 | 1 |
| II | PpeERF#010 | Prupe.1g464400 | 453 | 150 | 17240.2 | 9.42 | 2 |
| II | PpeERF#011 | Prupe.6g231500 | 501 | 166 | 17849.1 | 8.72 | 0 |
| II | PpeERF#012 | Prupe.1g546100 | 501 | 166 | 17890.7 | 5.62 | 0 |
| II | PpeERF#013 | Prupe.1g500900 | 726 | 241 | 26308.4 | 5.61 | 0 |
| II | PpeERF#014 | Prupe.7g194400 | 648 | 215 | 23980.2 | 5.16 | 0 |
| II | PpeERF#015 | Prupe.1g001700 | 552 | 183 | 20044.5 | 7.74 | 0 |
| III | PpeERF#016 | Prupe.5g090700 | 558 | 185 | 19627.0 | 4.89 | 0 |
| III | PpeERF#017 | Prupe.5g090600 | 537 | 178 | 19407.7 | 5.67 | 0 |
| III | PpeERF#018 | Prupe.5g090400 | 543 | 180 | 19547.0 | 5.50 | 0 |
| III | PpeERF#019 | Prupe.5g090300 | 543 | 180 | 19510.8 | 5.50 | 0 |
| III | PpeERF#020 | Prupe.5g090800 | 555 | 184 | 19705.9 | 4.64 | 0 |
| III | PpeERF#021 | Prupe.2g289600 | 696 | 231 | 24485.1 | 4.83 | 0 |
| III | PpeERF#022 | Prupe.7g116300 | 609 | 202 | 21536.7 | 4.91 | 0 |
| III | PpeERF#023 | Prupe.2g190200 | 609 | 202 | 22713.9 | 5.87 | 1 |
| III | PpeERF#024 | Prupe.2g123200 | 831 | 276 | 30098.8 | 4.94 | 0 |
| III | PpeERF#025 | Prupe.5g065300 | 762 | 253 | 28183.5 | 4.91 | 0 |
| III | PpeERF#026 | Prupe.1g448600 | 771 | 256 | 27960.9 | 5.11 | 0 |
| III | PpeERF#027 | Prupe.7g222700 | 789 | 262 | 28536.4 | 4.94 | 0 |
| III | PpeERF#028 | Prupe.3g062800 | 732 | 243 | 26436.2 | 6.67 | 1 |
| III | PpeERF#029 | Prupe.7g115900 | 606 | 201 | 22481.2 | 4.95 | 0 |
| III | PpeERF#030 | Prupe.2g289500 | 690 | 229 | 24976.9 | 5.14 | 7 |
| III | PpeERF#031 | Prupe.5g090500 | 723 | 240 | 27217.8 | 7.69 | 0 |
| III | PpeERF#032 | Prupe.5g090100 | 711 | 236 | 26352.7 | 5.17 | 0 |
| III | PpeERF#033 | Prupe.5g090000 | 693 | 230 | 25658.8 | 5.13 | 7 |
| III | PpeERF#034 | Prupe.5g090200 | 699 | 232 | 26157.5 | 5.03 | 1 |
| III | PpeERF#035 | Prupe.5g089900 | 693 | 230 | 25521.9 | 7.78 | 0 |
| III | PpeERF#036 | Prupe.1g545700 | 522 | 173 | 19078.3 | 5.44 | 0 |
| III | PpeERF#037 | Prupe.1g545400 | 600 | 199 | 22101.0 | 5.54 | 0 |
| IV | PpeERF#038 | Prupe.2g253000 | 1041 | 346 | 37474.1 | 6.52 | 0 |
| IV | PpeERF#039 | Prupe.6g284400 | 1554 | 517 | 58370.7 | 4.74 | 0 |
| IV | PpeERF#040 | Prupe.2g256900 | 1167 | 388 | 42473.1 | 4.64 | 3 |
| IV | PpeERF#041 | Prupe.3g223300 | 591 | 196 | 21305.5 | 8.32 | 0 |
| IV | PpeERF#042 | Prupe.7g066700 | 858 | 285 | 32054.9 | 5.73 | 0 |
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
| IV | PpeERF#043 | Prupe.6g354000 | 672 | 223 | 23337.9 | 8.50 | 0 |
| IV | PpeERF#044 | Prupe.1g372100 | 768 | 255 | 28137.6 | 6.55 | 0 |
| V | PpeERF#045 | Prupe.3g096000 | 639 | 212 | 24310.2 | 5.94 | 0 |
| V | PpeERF#046 | Prupe.1g390800 | 588 | 195 | 22413.4 | 6.66 | 6 |
| V | PpeERF#047 | Prupe.1g480400 | 672 | 223 | 24759.7 | 9.10 | 0 |
| V | PpeERF#048 | Prupe.7g243600 | 732 | 243 | 26570.6 | 7.02 | 0 |
| V | PpeERF#049 | Prupe.3g084600 | 567 | 188 | 20855.3 | 5.97 | 0 |
| V | PpeERF#050 | Prupe.7g004900 | 945 | 314 | 34636.3 | 5.44 | 0 |
| V | PpeERF#051 | Prupe.6g004400 | 867 | 288 | 31688.5 | 6.01 | 0 |
| V | PpeERF#052 | Prupe.3g263000 | 1275 | 424 | 46904.3 | 4.58 | 0 |
| V | PpeERF#053 | Prupe.5g136200 | 939 | 312 | 33152.5 | 6.13 | 0 |
| V | PpeERF#054 | Prupe.1g084800 | 936 | 311 | 33626.0 | 8.92 | 0 |
| V | PpeERF#055 | Prupe.5g213800 | 675 | 224 | 25367.4 | 8.71 | 0 |
| VI | PpeERF#056 | Prupe.5g114100 | 1002 | 333 | 37019.0 | 4.98 | 0 |
| VI | PpeERF#057 | Prupe.2g306400 | 1098 | 365 | 41098.9 | 4.75 | 2 |
| VI | PpeERF#058 | Prupe.2g183200 | 864 | 287 | 32269.0 | 4.95 | 0 |
| VI | PpeERF#059 | Prupe.3g019900 | 570 | 189 | 20614.4 | 9.37 | 0 |
| VI | PpeERF#060 | Prupe.7g060700 | 588 | 195 | 21933.2 | 10.37 | 2 |
| VI | PpeERF#061 | Prupe.6g039700 | 927 | 308 | 34031.9 | 5.49 | 2 |
| VI | PpeERF#062 | Prupe.5g220700 | 1311 | 436 | 49386.8 | 6.25 | 1 |
| VI | PpeERF#063 | Prupe.1g310100 | 1020 | 339 | 38021.2 | 4.92 | 1 |
| VII | PpeERF#064 | Prupe.8g264900 | 966 | 321 | 35682.6 | 5.68 | 4 |
| VII | PpeERF#065 | Prupe.3g032300 | 1149 | 382 | 42495.9 | 4.81 | 84 |
| VII | PpeERF#066 | Prupe.1g130300 | 870 | 289 | 31564.0 | 7.66 | 31 |
| VIII | PpeERF#067 | Prupe.3g094700 | 870 | 289 | 31781.0 | 9.56 | 0 |
| VIII | PpeERF#068 | Prupe.1g245500 | 1275 | 424 | 46297.0 | 7.73 | 0 |
| VIII | PpeERF#069 | Prupe.1g212700 | 831 | 276 | 29938.4 | 4.94 | 0 |
| VIII | PpeERF#070 | Prupe.1g139600 | 1092 | 363 | 39938.7 | 5.60 | 0 |
| VIII | PpeERF#071 | Prupe.4g176200 | 687 | 228 | 24245.0 | 9.62 | 2 |
| VIII | PpeERF#072 | Prupe.4g051400 | 795 | 264 | 28447.6 | 9.47 | 1 |
| VIII | PpeERF#073 | Prupe.8g230600 | 546 | 181 | 19951.0 | 9.99 | 0 |
| VIII | PpeERF#074 | Prupe.4g051200 | 519 | 172 | 18303.7 | 9.20 | 3 |
| VIII | PpeERF#075 | Prupe.4g222300 | 696 | 231 | 25022.8 | 7.62 | 20 |
| VIII | PpeERF#076 | Prupe.3g209100 | 708 | 235 | 25471.3 | 6.58 | 3 |
| IX | PpeERF#077 | Prupe.2g129700 | 672 | 223 | 24950.8 | 5.41 | 0 |
| IX | PpeERF#078 | Prupe.2g129500 | 651 | 216 | 23968.9 | 5.99 | 0 |
| IX | PpeERF#079 | Prupe.2g129600 | 714 | 237 | 25948.7 | 5.47 | 0 |
| IX | PpeERF#080 | Prupe.2g129300 | 816 | 271 | 29817.0 | 5.17 | 0 |
| IX | PpeERF#081 | Prupe.2g129400 | 621 | 206 | 22559.1 | 5.97 | 0 |
| IX | PpeERF#082 | Prupe.5g061800 | 846 | 281 | 30157.0 | 9.09 | 10 |
| IX | PpeERF#083 | Prupe.6g064700 | 2115 | 704 | 77761.3 | 9.77 | 15 |
| IX | PpeERF#084 | Prupe.2g272300 | 783 | 260 | 29210.8 | 6.53 | 3 |
| IX | PpeERF#085 | Prupe.5g062000 | 1059 | 352 | 39322.2 | 5.90 | 5 |
| Group name | Gene name | Locus name | ORF length (bp) | Size (aa) | Molecular weight (Da) | PI | EST hitting |
| IX | PpeERF#086 | Prupe.2g272400 | 945 | 314 | 35018.8 | 6.17 | 21 |
| IX | PpeERF#087 | Prupe.2g272500 | 726 | 241 | 27069.2 | 5.92 | 0 |
| IX | PpeERF#088 | Prupe.8g224700 | 540 | 179 | 20470.7 | 6.08 | 0 |
| IX | PpeERF#089 | Prupe.1g037800 | 492 | 163 | 18008.7 | 6.85 | 0 |
| IX | PpeERF#090 | Prupe.8g224800 | 465 | 154 | 17059.7 | 7.79 | 0 |
| IX | PpeERF#091 | Prupe.1g037700 | 417 | 138 | 15204.7 | 6.84 | 0 |
| IX | PpeERF#092 | Prupe.4g055600 | 795 | 264 | 29431.3 | 5.61 | 0 |
| IX | PpeERF#093 | Prupe.4g055500 | 564 | 187 | 20369.6 | 9.51 | 2 |
| IX | PpeERF#094 | Prupe.6g348700 | 744 | 247 | 28089.7 | 4.92 | 0 |
| IX | PpeERF#095 | Prupe.8g224600 | 759 | 252 | 28672.0 | 4.99 | 1 |
| IX | PpeERF#096 | Prupe.1g037900 | 699 | 232 | 25950.8 | 5.54 | 1 |
| X | PpeERF#097 | Prupe.8g125100 | 816 | 271 | 30318.2 | 8.16 | 0 |
| X | PpeERF#098 | Prupe.7g134100 | 801 | 266 | 29255.4 | 7.67 | 0 |
| X | PpeERF#099 | Prupe.5g141200 | 696 | 231 | 25763.5 | 9.21 | 0 |
| X | PpeERF#100 | Prupe.6g165700 | 1215 | 404 | 43314.9 | 7.07 | 2 |
| X | PpeERF#101 | Prupe.5g141300 | 816 | 271 | 30005.0 | 6.16 | 0 |
| X | PpeERF#102 | Prupe.1g214900 | 741 | 246 | 27203.7 | 6.23 | 1 |
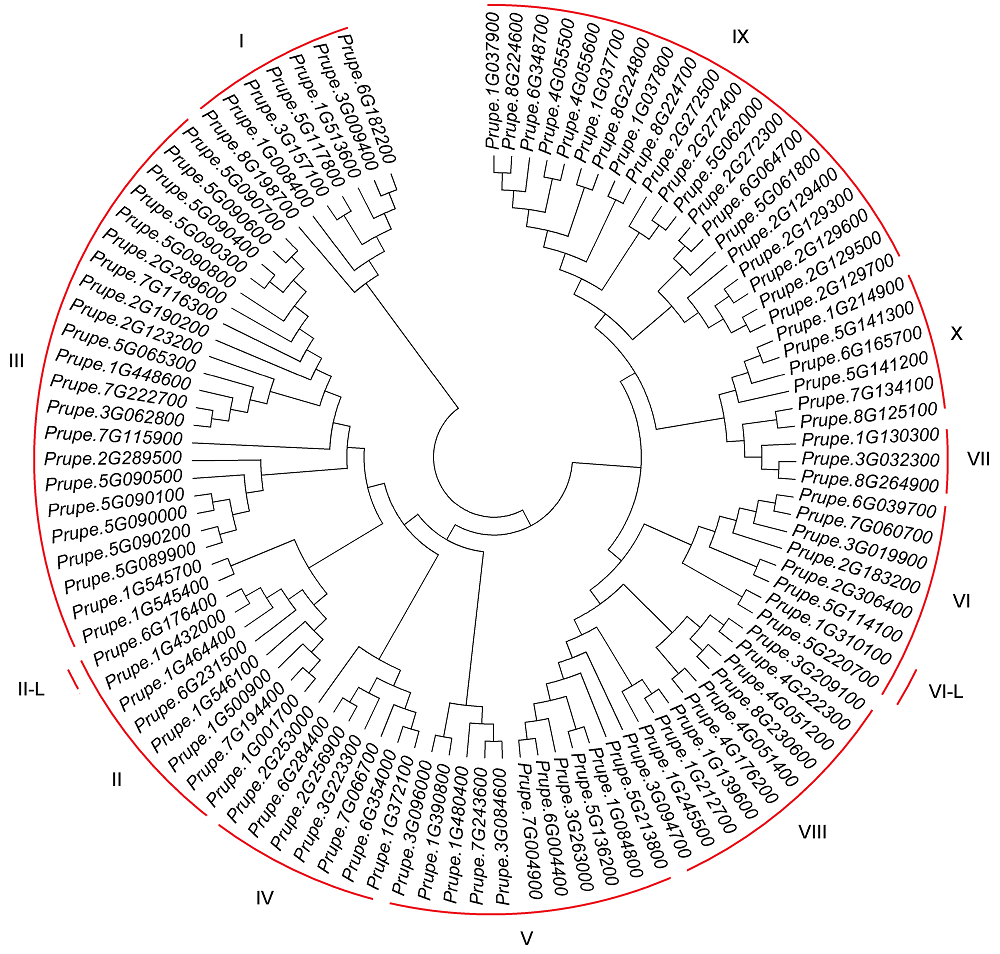
图1 桃ERF家族基因系统进化树I-X分别表示桃ERF家族的10个子家族; II-L: II-like; VI-L: VI-like
Figure 1 The phylogenetic tree of peach ERF family genesI-X indicate the 10 different groups of ERF family, respectively; II-L: II-like; VI-L: VI-like
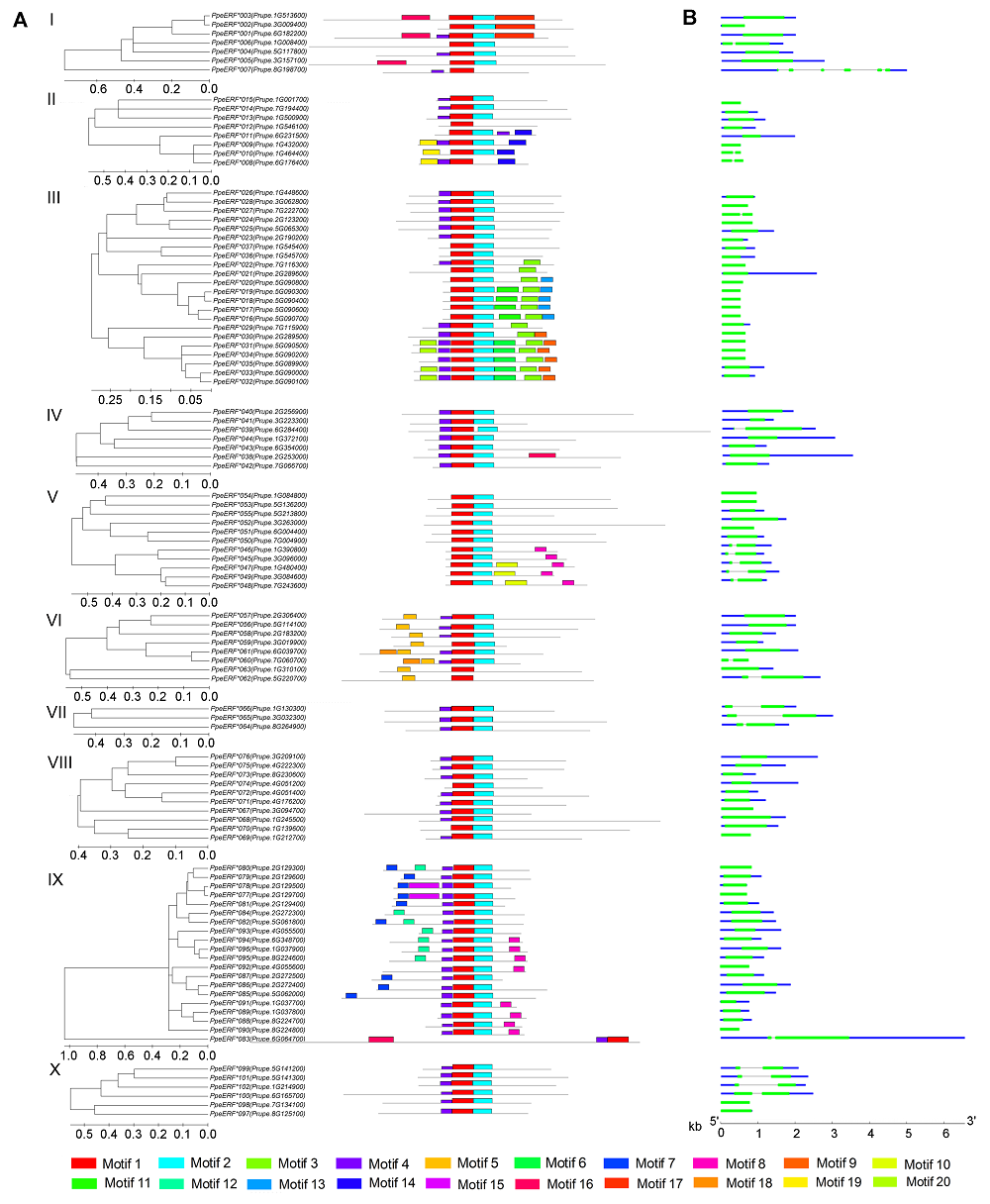
图3 桃ERF家族保守元件(A)和基因结构(B)I-X分别代表桃ERF家族的10个子家族。
Figure 3 Conserved motif (A) and structures (B) of ERF genes in peachI-X indicate the 10 different groups of ERF family, respectively.
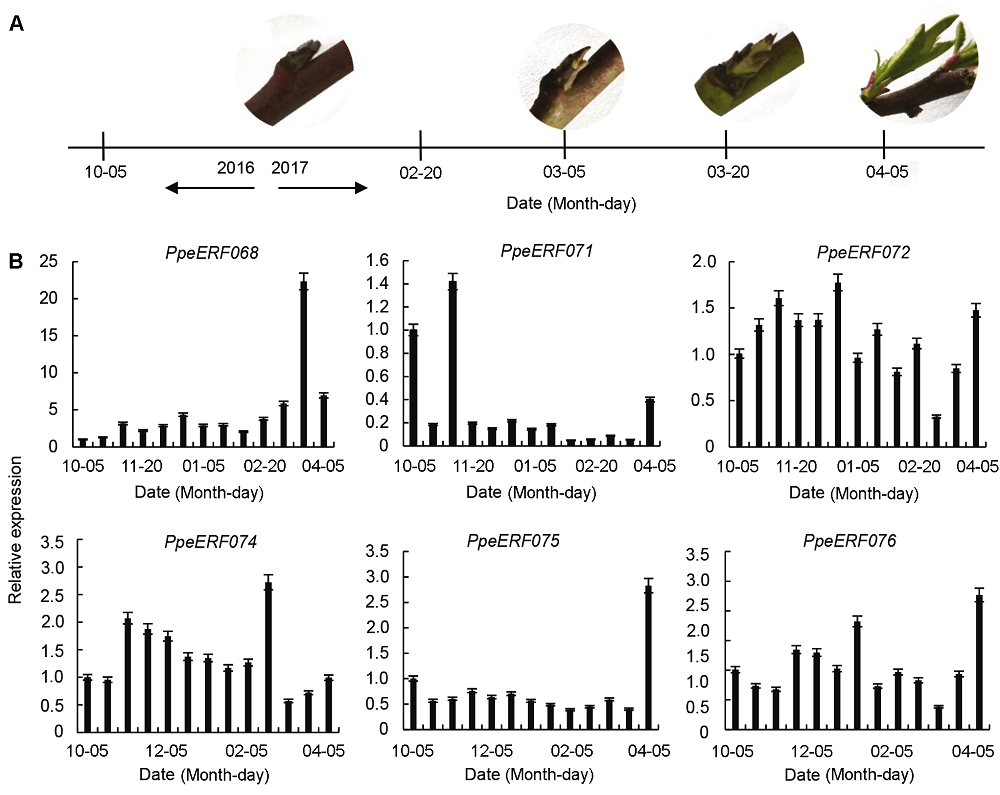
图4 自然条件下桃芽发育过程(A)及VIII子家族基因的表达(B)
Figure 4 Bud development process (A) and relative expression of VIII sub-family (B) in peach under natural condition
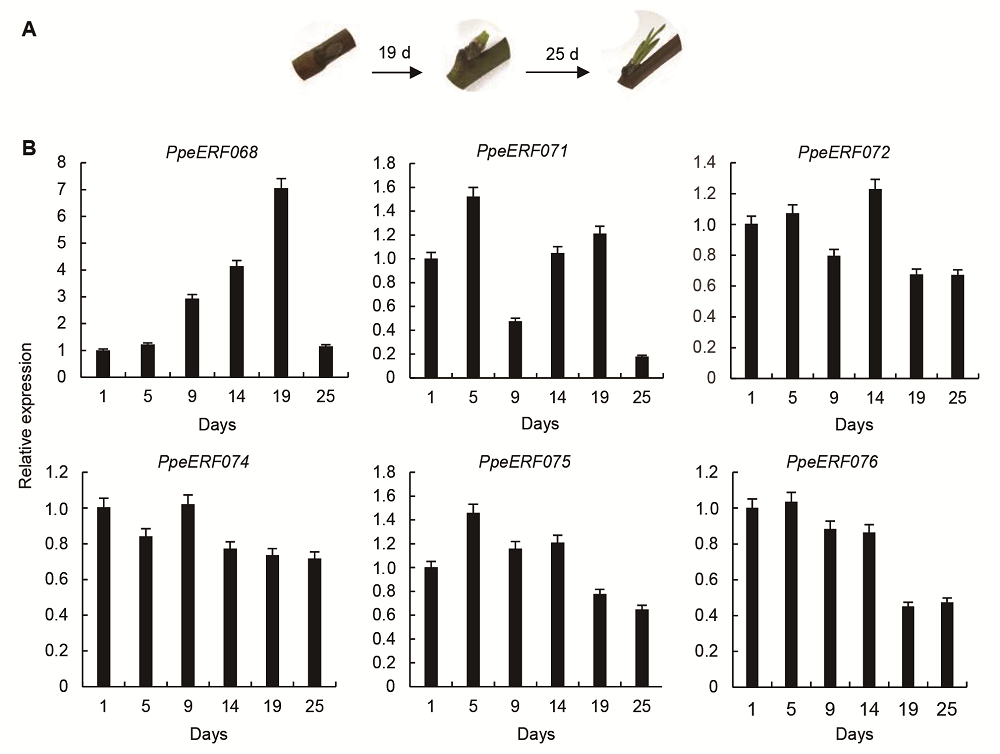
图5 光照培养箱中桃芽萌发过程(A)及VIII子家族基因的表达(B)
Figure 5 Bud development process (A) and relative expression of VIII sub-family genes (B) in peach in light culture incubation
| 1 | 段成国, 李宪利, 高东升, 刘焕芳, 李萌 (2004). 剥鳞和激素处理对大樱桃花芽休眠解除及内源激素变化的影响. 西北植物学报 24, 615-620. |
| 2 | 胡健兵 (2016). 全基因组发掘甜橙胁迫应答ERF类基因及其表达分析. 硕士论文. 武汉: 华中农业大学. pp. 27-33. |
| 3 | 孙明岳, 周君, 谭秋平, 付喜玲, 陈修德, 李玲, 高东升 (2016). 苹果bZIP转录因子家族生物信息学分析及其在休眠芽中的表达. 中国农业科学 49, 1325-1345. |
| 4 | 谭志一, 董毅敏, 高秀英, 房耀仁 (1985). 毛白杨冬芽休眠解除过程中脱落酸及赤霉素含量的变化. 植物学报 27, 381-386. |
| 5 | 翟莹, 杨晓杰, 孙天国, 赵艳, 余春粉, 王秀文 (2013). 大豆转录因子GmERF5的克隆、表达及功能分析. 植物学报 48, 498-506. |
| 6 | Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis.Plant Cell 16, 2463-2480. |
| 7 | Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis.Proc Natl Acad Sci USA 101, 4706-4711. |
| 8 | Carles CC, Fletcher JC (2003). Shoot apical meristem maintenance: the art of a dynamic balance.Trends Plant Sci 8, 394-401. |
| 9 | Chandler JW, Cole M, Jacobs B, Comelli P, Werr W (2011). Genetic integration of DORNRÖSCHEN and DORNRÖSCHEN-LIKE reveals hierarchical interactions in auxin signaling and patterning of the Arabidopsis apical embryo. Plant Mol Biol 75, 223-236. |
| 10 | Chandler JW, Werr W (2014). Arabidopsis floral phytomer development: auxin response relative to biphasic modes of organ initiation.J Exp Bot 65, 3097-3110. |
| 11 | Chen M, Tan QP, Sun MY, Li DM, Fu XL, Chen XD, Xiao W, Li L, Gao DS (2016). Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol Genet Genomics 291, 1319-1332. |
| 12 | Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt and cold-responsive gene expression. Plant J 33, 751-763. |
| 13 | Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155-168. |
| 14 | Ferguson BJ, Beveridge CA (2009). Roles for auxin, cytokinin, and strigolactone in regulating shoot branching.Plant Physiol 149, 1929-1944. |
| 15 | Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression.Plant Cell 12, 393-404. |
| 16 | Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang CM, He XH, Han Y, Martin GB (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis.Plant Cell 14, 817-831. |
| 17 | Hu YX, Wang YX, Liu XF, Li JY (2004). Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development.Cell Res 14, 8-15. |
| 18 | Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37, 720-729. |
| 19 | Moose SP, Sisco PH (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10, 3018-3027. |
| 20 | Nakano T, Suzuki K, Fujimura T, Shinshi H (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice.Plant Physiol 140, 411-432. |
| 21 | Pandey GK, Grant JJ, Cheong YH, Kim BG, Li LG, Luan S (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis.Plant Physiol 139, 1185-1193. |
| 22 | Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035-1046. |
| 23 | Savaldi-Goldstein S, Chory J (2008). Growth coordination and the shoot epidermis.Curr Opin Plant Biol 11, 42-48. |
| 24 | Sherif S, El-Sharkawy I, Paliyath G, Jayasankar S (2013). PpERF3b, a transcriptional repressor from peach, contribu- tes to disease susceptibility and side branching in EAR- dependent and -independent fashions.Plant Cell Rep 32, 1111-1124. |
| 25 | Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang PC, Zhu JK (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses.Plant Cell 17, 2384-2396. |
| 26 | Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K (2005). Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor.Plant Physiol 138, 675-685. |
| 27 | Xu W, Li F, Ling LZ, Liu AZ (2013). Genome-wide survey and expression profiles of the AP2/ERF family in castor bean(Ricinus communis L.). BMC Genomics 14, 785. |
| 28 | Xu ZS, Chen M, Li LC, Ma YZ (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement.J Integr Plant Biol 53, 570-585. |
| 29 | Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D (2004). The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis.Plant Phy- siol 136, 2862-2874. |
| 30 | Yordanov YS, Ma C, Strauss SH, Busov VB (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees.Proc Natl Acad Sci USA 111, 10001-10006. |
| 31 | Zhang GY, Chen M, Chen XP, Xu ZS, Guan S, Li LC, Li AL, Guo JM, Mao L, Ma YZ (2008). Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot 59, 4095-4107. |
| 32 | Zheng CL, Halaly T, Acheampong AK, Takebayashi Y, Jikumaru Y, Kamiya Y, Or E (2015). Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism.J Exp Bot 66, 1527-1542. |
| 33 | Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XY, Tian YS, Zhao W, Qiao YS, Zhang Z, Xiong AS, Yao QH (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun 371, 468-474. |
| [1] | 熊高明, 申国珍, 徐文婷, 谢宗强, 李跃林, 徐耀粘, 陈芳清, 李家湘. 中国低山丘陵热性常绿阔叶灌丛主要类型及群落特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 沈会涛, 俞筱押, 秦彦杰, 武爱彬. 太行山东麓核桃林碳氮磷化学计量及碳储量随林龄变化特征[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-. |
| [3] | 王燕玲, 招礼军, 朱栗琼, 莫若果, 林婷, 赵小雨. 广西天然红鳞蒲桃种群幼苗数量特征及动态分析[J]. 植物生态学报, 2023, 47(9): 1278-1286. |
| [4] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [5] | 管岳, 王妍欣, 褚佳瑶, 冯琳骄, 宋晓萌, 周龙. 新疆野扁桃种群年龄结构及动态分析[J]. 植物生态学报, 2023, 47(7): 967-977. |
| [6] | 闫涵, 马松梅, 魏博, 张宏祥, 张丹. 孑遗灌木长柄扁桃的历史分布格局及其环境驱动力[J]. 植物生态学报, 2022, 46(7): 766-774. |
| [7] | 李晨, 刘建廷, 樊永信, 赵雪惠, 肖伟, 陈修德, 付喜玲, 李玲, 李冬梅. UV-B对设施桃叶片光合功能及叶绿体超微结构的影响[J]. 植物学报, 2022, 57(4): 434-443. |
| [8] | 周亮, 杨君珑, 杨虎, 窦建德, 黄维, 李小伟. 宁夏蒙古扁桃群落特征与分类[J]. 植物生态学报, 2022, 46(2): 243-248. |
| [9] | 王春成, 张云玲, 马松梅, 黄刚, 张丹, 闫涵. 中国扁桃亚属四种野生扁桃的系统发育与物种分化[J]. 植物生态学报, 2021, 45(9): 987-995. |
| [10] | 贾晓东,许梦洋,莫正海,宣继萍,翟敏,郭忠仁. 薄壳山核桃酚类代谢物研究进展[J]. 植物学报, 2020, 55(1): 106-119. |
| [11] | 唐丽丽, 张梅, 赵香林, 康慕谊, 刘鸿雁, 高贤明, 杨彤, 郑璞帆, 石福臣. 华北地区胡桃楸林分布规律及群落构建机制分析[J]. 植物生态学报, 2019, 43(9): 753-761. |
| [12] | 张淑辉,王红,王文茹,吴雪莲,肖元松,彭福田. 蔗糖对桃幼苗生长发育及其SnRK1酶活性的影响[J]. 植物学报, 2019, 54(6): 744-752. |
| [13] | 郜怀峰,张亚飞,王国栋,孙希武,贺月,彭福田,肖元松. 钼在桃树干旱胁迫响应中的作用解析[J]. 植物学报, 2019, 54(2): 227-236. |
| [14] | 张媚, 林马水, 曹秀秀, 赵树民, 蒋达青, 王冰璇, 汪石莹, 樊炎迪, 郭明, 林海萍. 不同经营模式山核桃林地土壤pH值、养分与细菌多样性的差异[J]. 生物多样性, 2018, 26(6): 611-619. |
| [15] | 周乃富, 张俊佩, 刘昊, 查巍巍, 裴东. 木本植物非均质化组织石蜡切片制作方法[J]. 植物学报, 2018, 53(5): 653-660. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||