

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (2): 184-193.DOI: 10.11983/CBB15080 cstr: 32102.14.CBB15080
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Kaijian Lei1,2, Jing Ren1, Yuanyuan Zhu1, Guoyong An1,*( )
)
Received:2015-05-18
Accepted:2015-12-13
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: Kaijian Lei, Jing Ren, Yuanyuan Zhu, Guoyong An. SPL1 is Involved in the Regulation of Rhizosphere Acidification Reaction Under Low Phosphate Condition in Arabidopsis[J]. Chinese Bulletin of Botany, 2016, 51(2): 184-193.
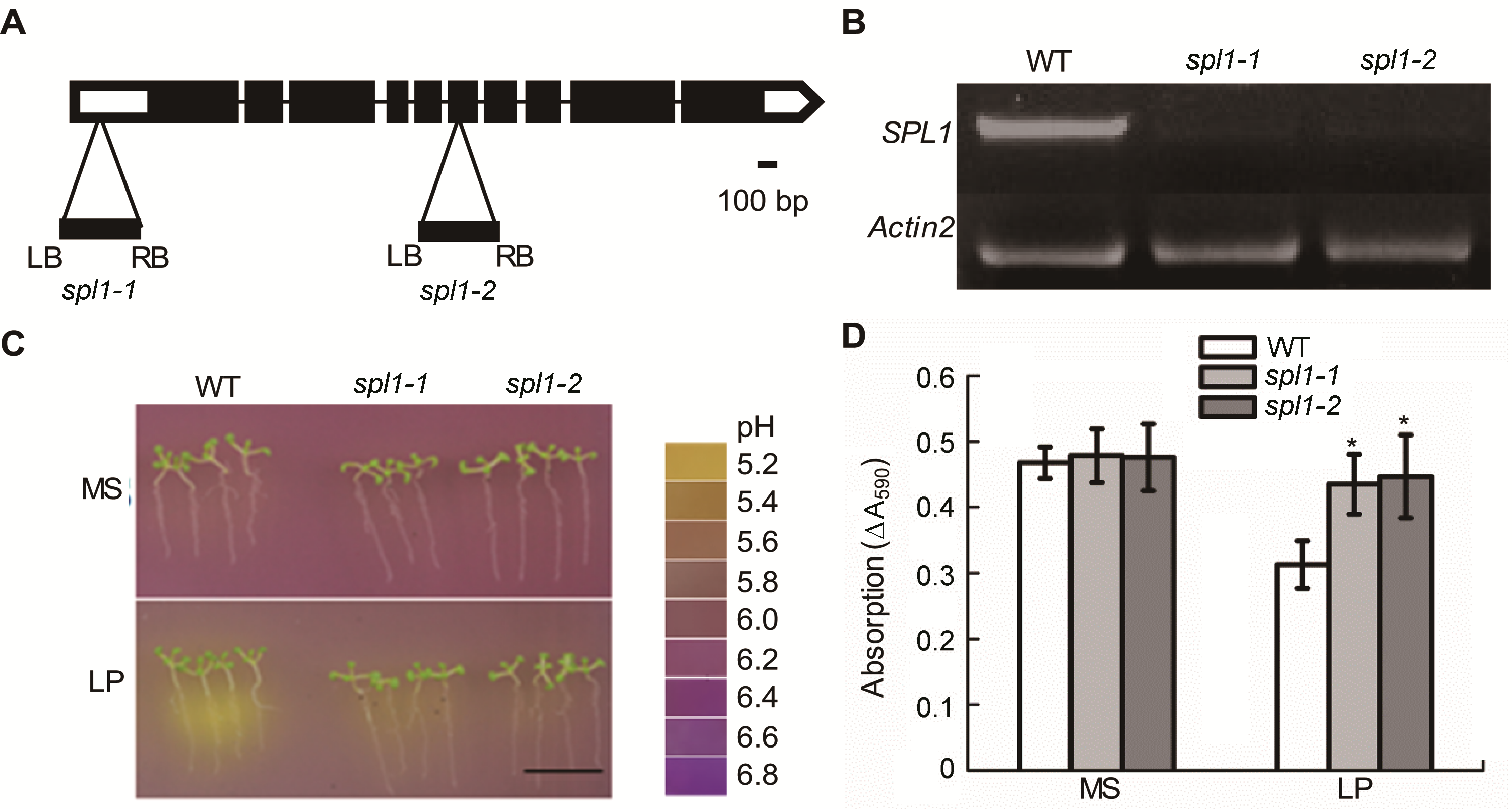
Figure 1 Arabidopsis thaliana spl1 mutants show the phenotype of rhizosphere acidification deficiency and decreased acidi- fication capacity (A) T-DNA insertion sites in spl1-1 and spl1-2; (B) Reverse transcriptase-PCR analysis of SPL1 expression; (C) The rhizosphere acidification reaction of WT and spl1 mutants under MS and low phosphate (LP) conditions (12.5 μmo∙L−1 H2PO4−) (Bar=1 cm); (D) The acidification capacity of WT and spl1 mutants. Asterisk represent statistically differences compared with the wild type (P<0.05).
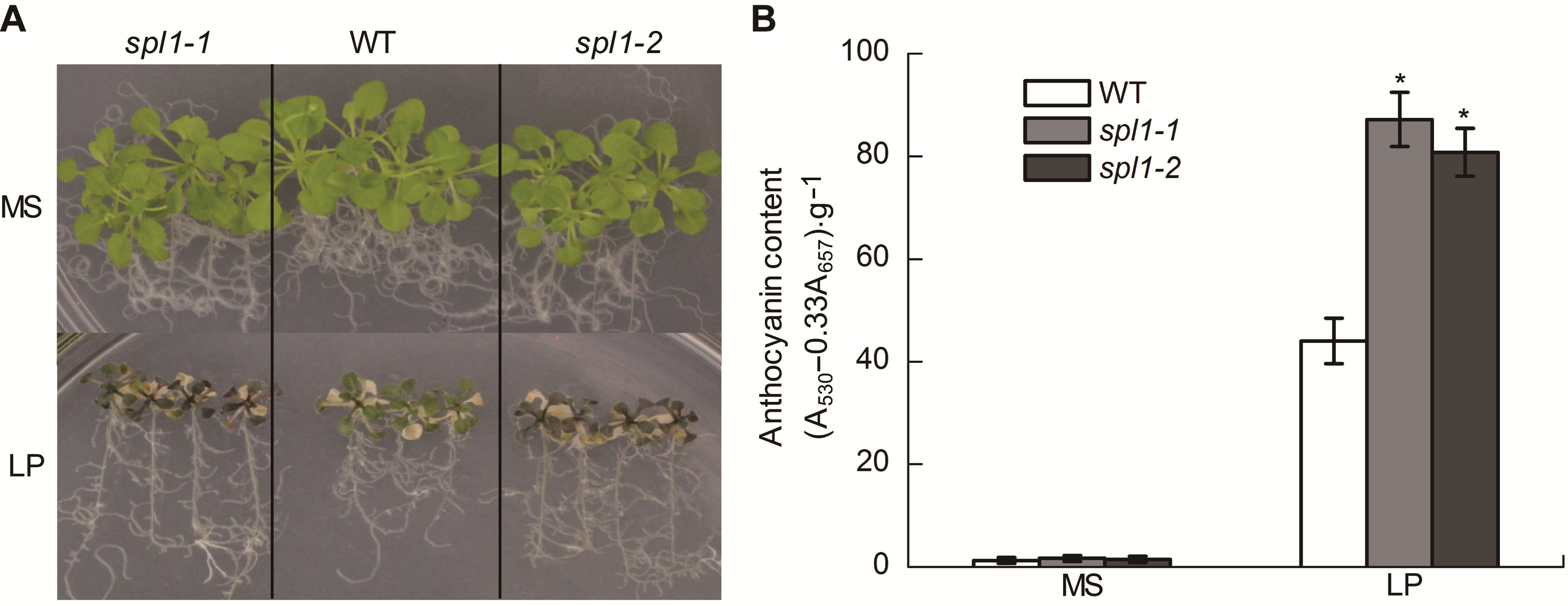
Figure 2 The anthocyanin accumulation in Arabidopsis thaliana spl1-1 and spl1-2 under low Pi condition (A) The anthocyanin accumulation in wild type (WT), spl1-1, and spl1-2 plants during Pi sufficient and Pi deprivation; (B) Anthocyanin content was determined in WT and spl1 plants grown with normal and low Pi (LP) on the twentieth day of Pi starvation. Asterisk represents statistically differences compared with the wild type (P<0.05).
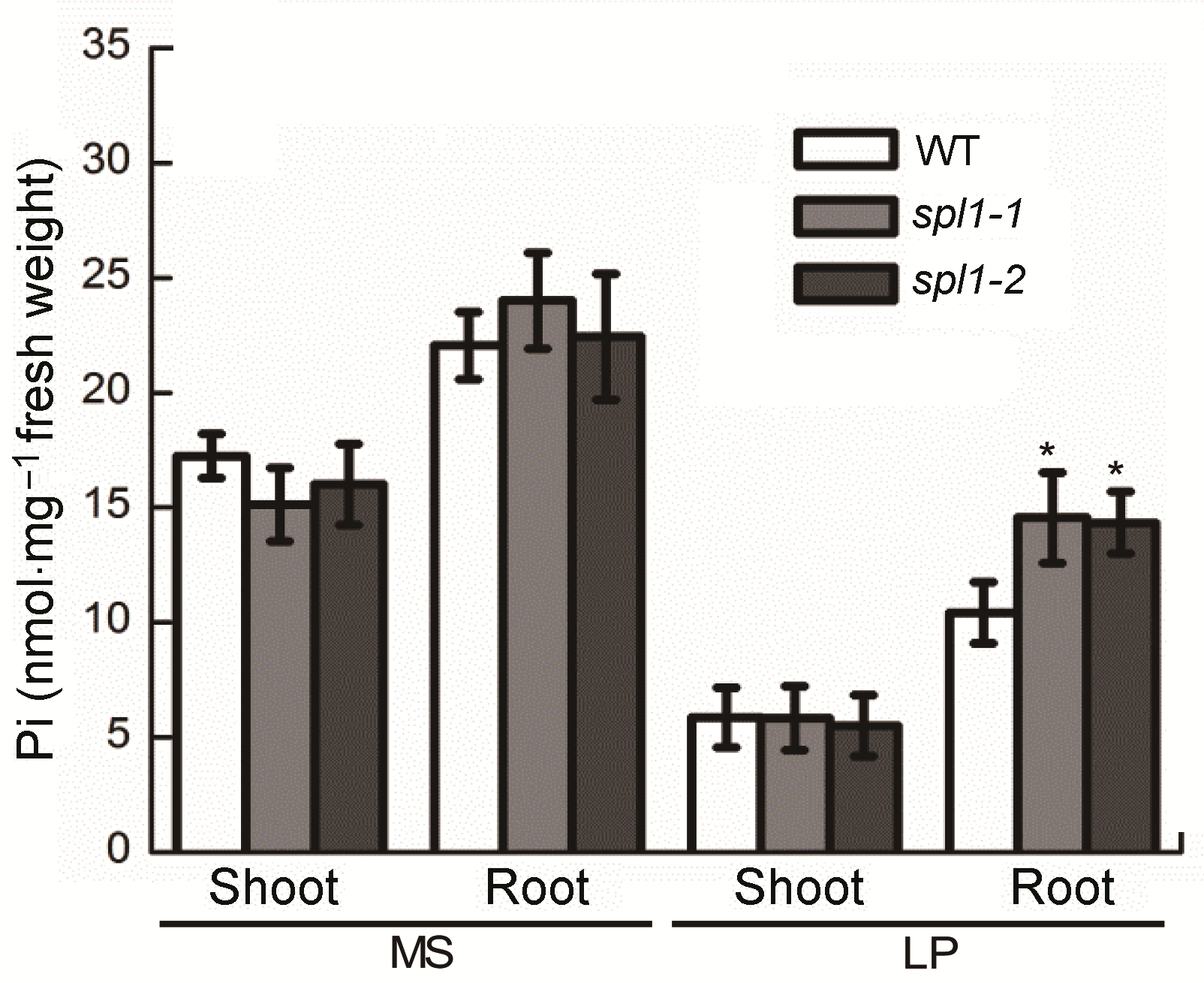
Figure 3 The Pi content analysis of Arabidopsis thaliana spl1 mutants Seven-day-old seedlings were transferred to MS or low Pi (LP) medium for 14 d and then harvested for Pi content analy- sis. Asterisk represents statistically differences compared with the wild type (P<0.05).
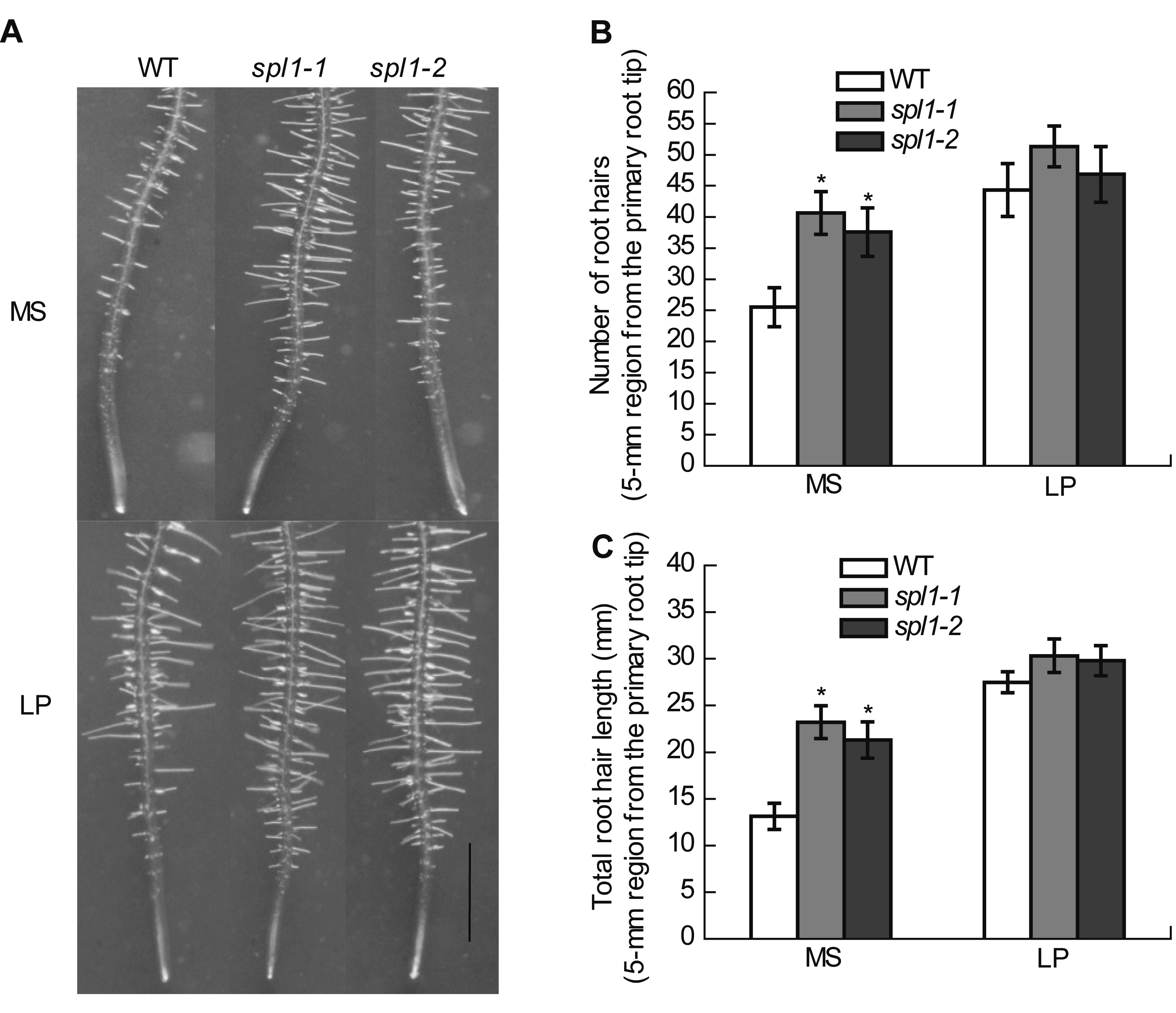
Figure 4 SPL1 alters root hair architecture (A) Images of root tips with intact root hair from WT, spl1-1 and spl1-2 plants (Bar=1 mm); (B), (C) Number of root hair (B) and root hair length (C) under MS and low Pi (LP) condition. *Data significantly different from the corresponding controls are indicated (P<0.05).
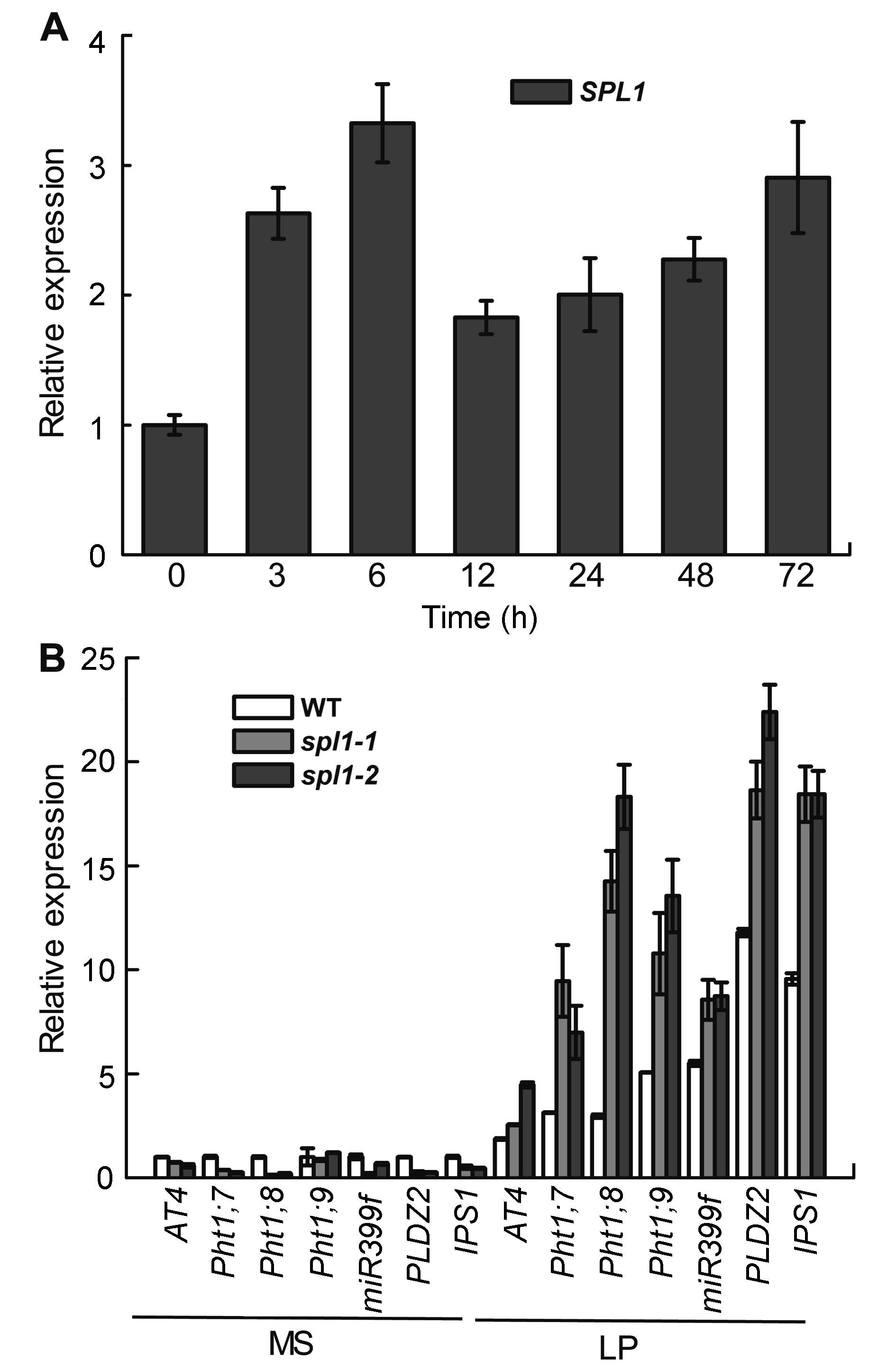
Figure 5 Expression analyses of the SPL1 gene (A) in Arabidopsis seedlings in response to low phosphate (LP) stress and expression pattern of Pi starvation induced genes in spl1 mutants (B)
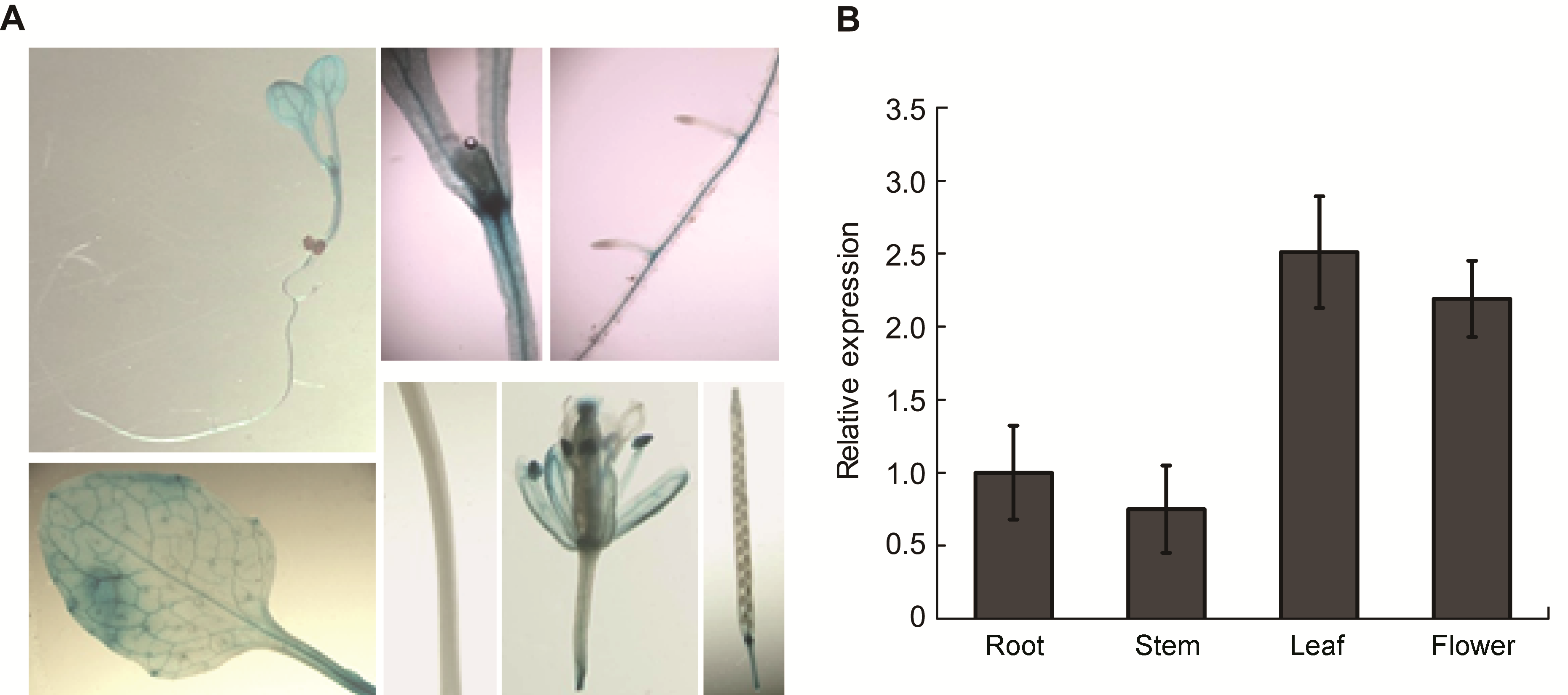
Figure 6 Tissue-specific expression of SPL1 (A) Histochemical localization of GUS activity directed by SPL1::GUS fusions in transgenic Arabidopsis; (B) qRT-PCR showed the relative abundant of SPL1 gene in different tissues, including root, stem, leaf and flower
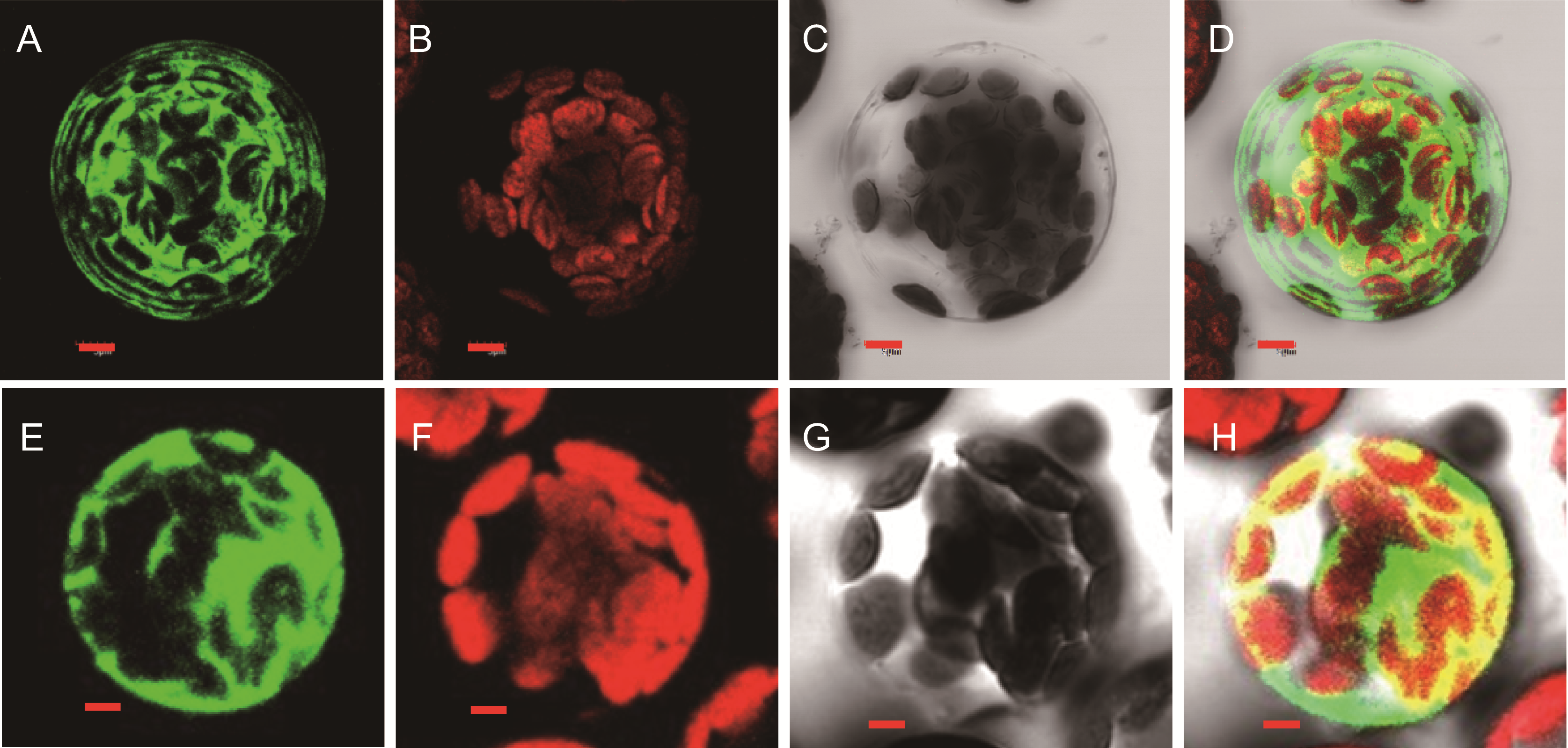
Figure 7 Localization of GFP signals from pHBT empty vector (A−D) and SPL1 (E−H) fused with GFP (A), (E) Fluorescence images under confocal microscopy; (B), (F) Chloroplast fluorescence; (C), (G) Bright-field images of the cells; (D), (H) Merged fluorescence. Bar=5 µm
| [1] | 雷凯健, 安国勇 (2014). 植物miRNA介导磷信号转导的研究进展. 植物生理学报 50, 1071-1078. |
| [2] | 张健, 徐金相, 孔英珍, 纪振动, 王兴春, 安丰英, 李超, 孙加强, 张素芝, 杨晓辉, 牟金叶, 刘新仿, 李家洋, 薛勇彪, 左建儒 (2005). 化学诱导激活型拟南芥突变体库的构建及分析. 遗传学报 32, 1082-1088. |
| [3] | Ames BN (1966). Assay of inorganic phosphate, total phosphate and phosphatase. Methods Enzymol 8, 115-118. |
| [4] | Cordell D, Dranger JO, White S (2009). The story of phosphorus: global food security and food for thought. Global Environ Change 19, 292-305. |
| [5] |
Cruz-Ramirez A, Oropeza-Aburto A, Razo-Hernandez F, Ramirez-Chavez E, Herrera-Estrella L (2006). Phos- pholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103, 6765-6770.
DOI PMID |
| [6] | Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80, 1108-1117. |
| [7] |
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39, 1033-1037.
DOI PMID |
| [8] |
Gomes-Junior RA, Moldes CA, Delite FS, Pompeu GB, Gratão PL, Mazzafera P, Lea PJ, Azevedo RA (2006). Antioxidant metabolism of coffee cell suspension cultures in response to cadmium. Chemosphere 65, 1330-1337.
DOI PMID |
| [9] | Gou JY, Felippes F, Liu CJ, Weigel D, Wang JW (2011). Negative regulation of anthocyanin biosynthesis in Ara- bidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512-1522. |
| [10] |
Hinsinger P, Gobran GR, Gregory PJ, Wenzel WW (2005). Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytol 168, 293-303.
DOI PMID |
| [11] |
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009). Uncovering small RNA-media- ted responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151, 2120-2132.
DOI PMID |
| [12] |
Jefferson RA, Kavanagh TA, Bevan MW (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6, 3901-3907.
DOI PMID |
| [13] |
Khorassani R, Hettwer U, Ratzinger A, Steingrobe B, Karlovsky P, Claassen N (2011). Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol 11, 121-128.
DOI PMID |
| [14] | Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduc- tion. Plant Cell 15, 2399-2407. |
| [15] | Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient tem- perature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol 159, 461-478. |
| [16] |
Lapis-Gaza HR, Jost R, Finnegan PM (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14, 334.
DOI PMID |
| [17] | Lei KJ, Lin YM, Ren J, Bai L, Miao YC, An GY, Song CP (2016). Modulation of the phosphate-deficient responses by the microRNA156 and its targeted SQUAMOSA PRO- MOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol 57, 192-203. |
| [18] | Lei KJ, Xie JY, Zhu YY, Song CP, An GY (2015). Screen- ing and analysis of rhizosphere acidification deficiency mutants in Arabidopsis thaliana under low phosphorus. Soil Sci Plant Nutr 61, 493-500. |
| [19] | Li M, Shinano T, Tadano T (1997). Distribution of exu- dates of lupin roots in the rhizosphere under phosphor- rus deficient conditions. Soil Sci Plant Nutr 43, 237-245. |
| [20] | Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147, 732-746. |
| [21] |
López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003). The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6, 280-287.
DOI PMID |
| [22] | Neumann G, Römheld V (1999). Root excretion of carbo- xylic acids and protons in phosphorus-deficient plants. Plant Soil 211, 121-130. |
| [23] | Raghothama KG (1999). Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50, 665-693. |
| [24] |
Rausch C, Bucher M (2002). Molecular mechanisms of phosphate transport in plants. Planta 216, 23-37.
DOI PMID |
| [25] |
Rouached H, Arpat AB, Poirier Y (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3, 288-299.
DOI PMID |
| [26] | Santi S, Schmidt W (2009). Dissecting iron deficiency- induced proton extrusion in Arabidopsis roots. New Phytol 183, 1072-1084. |
| [27] | Shin H, Shin HS, Chen R, Harrison MJ (2006). Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45, 712-726. |
| [28] |
Sternberg D, Mandels GR (1979). Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol 139, 761-769.
DOI PMID |
| [29] | Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcript- tion factors. Plant Cell 26, 1792-1807. |
| [30] | Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acqui- sition in Arabidopsis. Plant Physiol 67, 1579-1591. |
| [31] |
Ticconi CA, Abel S (2004). Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9, 548-555.
PMID |
| [32] | Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P (2003). SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15, 1009-1019. |
| [33] |
Vance CP, Uhde-Stone C, Allan DL (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157, 423-447.
DOI PMID |
| [34] | Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008). Dual effects of miR156-targeted SPL genes and CYP- 78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231-1243. |
| [35] |
Watt M, Evans JR (1999). Proteoid roots. Physiology and development. Plant Physiol 121, 317-323.
PMID |
| [36] |
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750-759.
DOI PMID |
| [37] | Xing SP, Salinas M, Höhmann S, Berndtgen R, Huijser P (2010). MiR156-targeted and non targeted SBP-box trans- cription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935-3950. |
| [38] |
Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009). SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 is a central regulator for copper homeo- stasis in Arabidopsis. Plant Cell 21, 347-361.
DOI PMID |
| [39] | Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTERBIN- DING-LIKE transcription factors. Plant Cell 24, 3320-3332. |
| [40] |
Zhang Y, Schwarz S, Saedler H, Huijser P (2007). SPL8, a local regulator in a subset of gibberellins mediated developmental processes in Arabidopsis. Plant Mol Biol 63, 429-439.
DOI PMID |
| [41] |
Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, Li X, Zhang Y (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107, 18220-18225.
DOI PMID |
| [1] | Yuhan Liu, Qijiang Cao, Shihan Zhang, Yihui Li, Jing Wang, Xiaomeng Tan, Xiaoru Liu, Xianling Wang. Mechanism of AtFTCD-L in Root Response to Soil Compaction [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Yanjun Jing, Rongcheng Lin. Blue Light Receptor CRY2 Transforms into a ‘dark dancer’ [J]. Chinese Bulletin of Botany, 2024, 59(6): 878-882. |
| [3] | Yan Luo, Qiyuan Liu, Yuanbing Lü, Yue Wu, Yaoyu Tian, Tian An, Zhenhua Li. Photothermal Sensitivity of Phytochrome Mutants During Seed Germination in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2024, 59(5): 752-762. |
| [4] | Yanxiao Chen, Yaping Li, Jinjun Zhou, Lixia Xie, Yongbin Peng, Wei Sun, Yanan He, onghui Jiang, Zenglan Wang, Chongke Zheng, Xianzhi Xie. Effect of Amino Acid Point Mutations on the Structure and Function of Phytochrome B in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2024, 59(3): 481-494. |
| [5] | Jixuan Yang, Xuefei Wang, Hongya Gu. Genetic Basis of Flowering Time Variations in Tibetan Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2024, 59(3): 373-382. |
| [6] | Gang Wang, Ertao Wang. The Broad-spectrum Innate Resistance Against Clubroot Disease Conferred by WeiTsing is Mechanistically Revealed [J]. Chinese Bulletin of Botany, 2023, 58(3): 356-358. |
| [7] | Yongguang Li, Hui Ren, Yingjie Zhang, Ruining Li, Hao Ai, Xianzhong Huang. Analysis of the molecular evolution of the PEBP gene family in cruciferous plants [J]. Biodiv Sci, 2022, 30(6): 21545-. |
| [8] | Yang Yongqing, Guo Yan. Analysis of the pH Sensing Mechanism of Plant Apoplasts [J]. Chinese Bulletin of Botany, 2022, 57(4): 409-411. |
| [9] | Tiantian Zhi, Zhou Zhou, Chengyun Han, Chunmei Ren. PAD4 Mutation Accelerating Programmed Cell Death in Arabidopsis thaliana Tyrosine Degradation Deficient Mutant sscd1 [J]. Chinese Bulletin of Botany, 2022, 57(3): 288-298. |
| [10] | Yanyan Li, Yanhua Qi. Advances in Biological Functions of Aux/IAA Gene Family in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 30-41. |
| [11] | Qiuxin Li, Wei Chi, Daili Ji. Research Progress of CURT1 on Regulating Thylakoid Membrane Curvature [J]. Chinese Bulletin of Botany, 2021, 56(4): 462-469. |
| [12] | Yongmei Che, Yanjun Sun, Songchong Lu, Lixia Hou, Xinxin Fan, Xin Liu. AtMYB77 Involves in Lateral Root Development via Regulating Nitric Oxide Biosynthesis under Drought Stress in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2021, 56(4): 404-413. |
| [13] | Ting Wang, Huanhuan Yang, Hongwei Zhao, Josef Voglmeir, Li Liu. Changes of Protein N-glycosylation in the Growth of Arabidopsis thaliana and Effects of Enzymatic Deglycosylation on Root Development [J]. Chinese Bulletin of Botany, 2021, 56(3): 262-274. |
| [14] | Yuqing Lin, Yanhua Qi. Advances in Auxin Efflux Carrier PIN Proteins [J]. Chinese Bulletin of Botany, 2021, 56(2): 151-165. |
| [15] | Long Ma, Guilin Li, Shipeng Li, Su Jiang. An Improved Protocol for Whole Mount Clearing of Plant Root Tip [J]. Chinese Bulletin of Botany, 2020, 55(5): 596-604. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||