

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (5): 520-532.DOI: 10.11983/CBB21119 cstr: 32102.14.CBB21119
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Jiangyuan Shang, Yan Chun, Xueyong Li*( )
)
Received:2021-07-21
Accepted:2021-09-16
Online:2021-09-01
Published:2021-09-16
Contact:
Xueyong Li
Jiangyuan Shang, Yan Chun, Xueyong Li. Map-based Cloning and Natural Variation Analysis of the PAL3 Gene Controlling Panicle Length in Rice[J]. Chinese Bulletin of Botany, 2021, 56(5): 520-532.
| Primer name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| R11-3 | AGAGAGACATCCGGAGACAA | TAAGACGAAAGGTCAAACGT |
| R11-5 | GTGCTAACGTTTCGTCTAAC | AATAGCCTTCGGTGGTCTCA |
| R11-10 | GTTCGTAATGTGGGCGTCTT | TGGGCACTCTTCTCACACTG |
| R11-12 | CAATCTTGCTCTACTAGCTAGTG | GTGGCAACTAACAGATTAGATG |
| M1 | GCAGTATATATTCGGCGGCG | GCCGTCGCCATATAGCTG |
| M2 | GAGCCTCTCCTACTGTGCTA | AGAGCCCTCAGTTCCTCAAT |
| M3 | GCTGACTACAGTAAGATCATGC | AGACAAACGGTCAAACATGT |
| M4 | AAGGATCCAAGCTAGCCTCC | CCTGACAGCAAGCGAGAGAT |
| M5 | CTTCAGCAAGTGAACTACGA | CCTAAACTAGCACGGATCATAGC |
| M6 | TGTGAGGTTTAGGTTCTCGGA | TGAATAGAGATGCGGTCCAAC |
| M7 | GGATTCGGCCACTGGTTGTT | GAATGTACTCGGATAAACCC |
Table 1 Primers used for mapping
| Primer name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| R11-3 | AGAGAGACATCCGGAGACAA | TAAGACGAAAGGTCAAACGT |
| R11-5 | GTGCTAACGTTTCGTCTAAC | AATAGCCTTCGGTGGTCTCA |
| R11-10 | GTTCGTAATGTGGGCGTCTT | TGGGCACTCTTCTCACACTG |
| R11-12 | CAATCTTGCTCTACTAGCTAGTG | GTGGCAACTAACAGATTAGATG |
| M1 | GCAGTATATATTCGGCGGCG | GCCGTCGCCATATAGCTG |
| M2 | GAGCCTCTCCTACTGTGCTA | AGAGCCCTCAGTTCCTCAAT |
| M3 | GCTGACTACAGTAAGATCATGC | AGACAAACGGTCAAACATGT |
| M4 | AAGGATCCAAGCTAGCCTCC | CCTGACAGCAAGCGAGAGAT |
| M5 | CTTCAGCAAGTGAACTACGA | CCTAAACTAGCACGGATCATAGC |
| M6 | TGTGAGGTTTAGGTTCTCGGA | TGAATAGAGATGCGGTCCAAC |
| M7 | GGATTCGGCCACTGGTTGTT | GAATGTACTCGGATAAACCC |
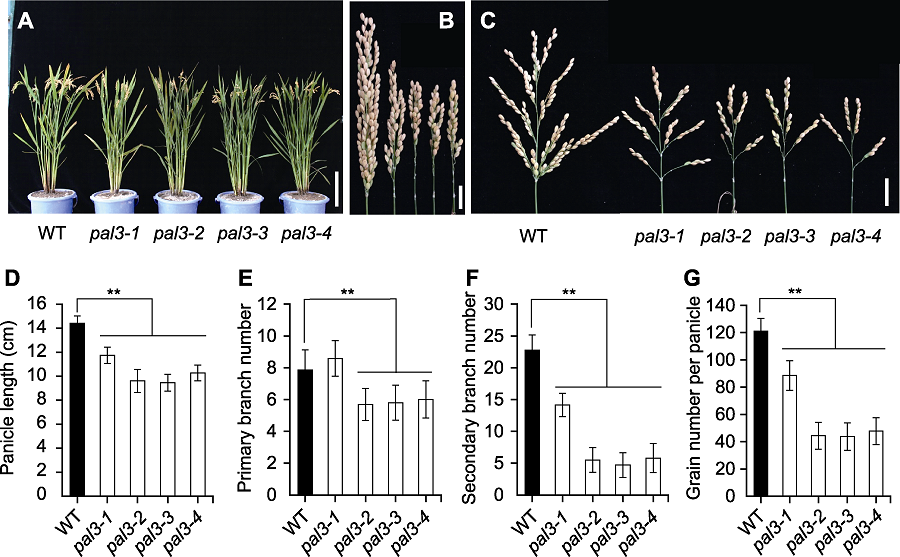
Figure 1 Comparison of the rice phenotype between wild type and the pal3 mutants (A) Gross morphology of wild type and the pal3 mutants at mature stage (Bar=20 cm); (B) Closed panicle of wild type and the pal3 mutants (Bar=2 cm); (C) Spread panicles of wild type and the pal3 mutants (right) (Bar=2 cm); (D)-(G) Statistic analysis of panicle length (D), primary branch number (E), secondary branch number (F) and grain number per panicle (G) of wild type and the pal3 mutants. WT: Wild type. Data in figure are means±SD (n=20). ** indicate the significant differences at P<0.01 level by Students’t test.
| Hybrid combi- nations | Phenotype of F1 | F2 population | χ23:1 | ||
|---|---|---|---|---|---|
| Wild-type plant number | Mutant-phenotypic plant number | Total number | |||
| pal3-1 × WT | WT | 190 | 60 | 250 | 0.13 |
| pal3-2 × WT | WT | 196 | 68 | 264 | 0.08 |
| pal3-3 × WT | WT | 180 | 63 | 243 | 0.11 |
| pal3-4 × WT | WT | 201 | 69 | 270 | 0.04 |
Table 2 Genetic analysis of the pal3-1, pal3-2, pal3-3 and pal3-4 rice mutants
| Hybrid combi- nations | Phenotype of F1 | F2 population | χ23:1 | ||
|---|---|---|---|---|---|
| Wild-type plant number | Mutant-phenotypic plant number | Total number | |||
| pal3-1 × WT | WT | 190 | 60 | 250 | 0.13 |
| pal3-2 × WT | WT | 196 | 68 | 264 | 0.08 |
| pal3-3 × WT | WT | 180 | 63 | 243 | 0.11 |
| pal3-4 × WT | WT | 201 | 69 | 270 | 0.04 |
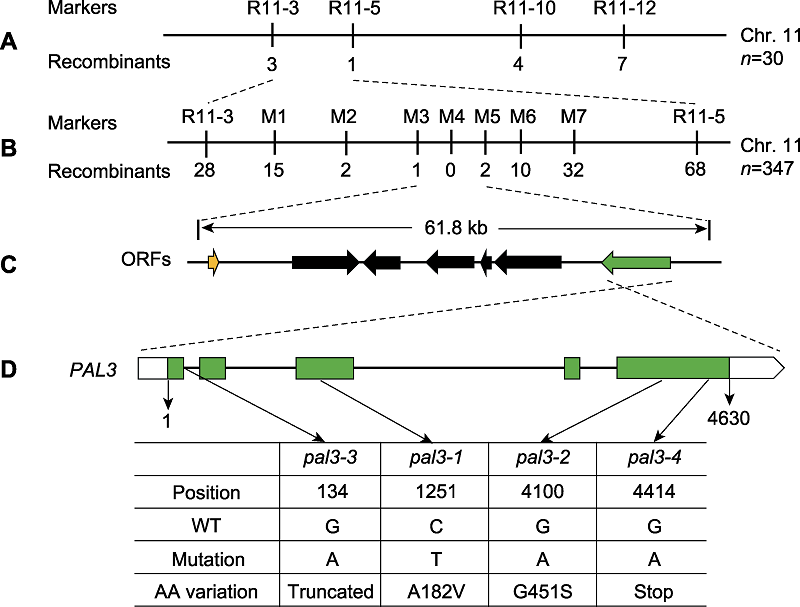
Figure 2 Map-based cloning of the PAL3 gene (A) The PAL3 gene was mapped to a region between InDel markers R11-3 and R11-5 on chromosome 11; (B), (C) Fine-mapping of PAL3. The PAL3 gene was further delimited to the region between markers M3 and M5, which contains seven open reading frames (ORFs). The number of recombinants is indicated beneath the marker positions; (D) Schematic structure of the PAL3 gene and the mutation sites of pal3 mutants. WT: Wild type
| Gene ID | Annotation |
|---|---|
| LOC_Os11g12680 | Expressed protein |
| LOC_Os11g12690 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12700 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12710 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12720 | Retrotransposon, putative, centromere-specific |
| LOC_Os11g12730 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| LOC_Os11g12740 | Peptide transporter PTR2, putative, expressed |
Table 3 Annotations of candidate genes in the fine-mapping region
| Gene ID | Annotation |
|---|---|
| LOC_Os11g12680 | Expressed protein |
| LOC_Os11g12690 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12700 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12710 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12720 | Retrotransposon, putative, centromere-specific |
| LOC_Os11g12730 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| LOC_Os11g12740 | Peptide transporter PTR2, putative, expressed |
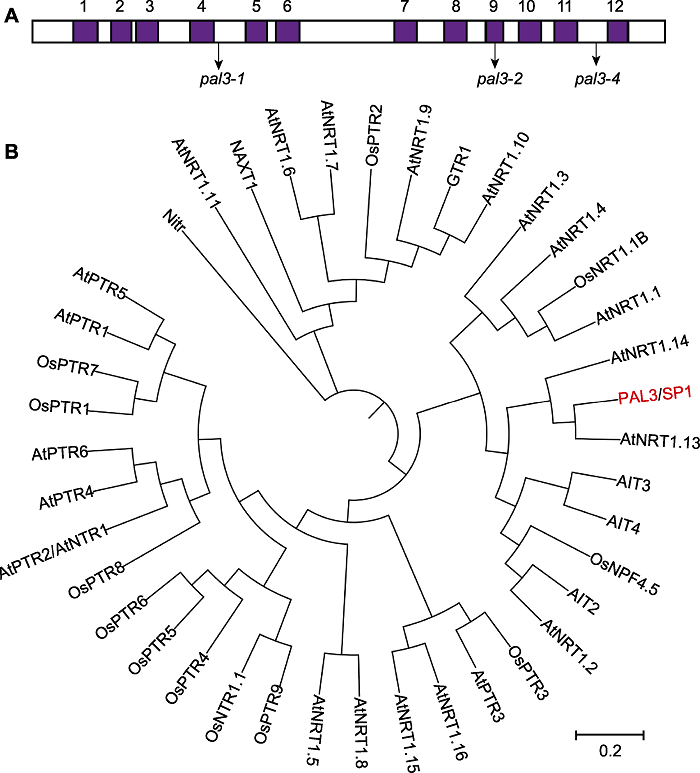
Figure 3 Transmembrane regions and phylogenetic analysis of PAL3 (A) 12 transmembrane domains of PAL3 and the mutation sites of the pal3-1, pal3-2 and pal3-4 mutants; (B) Phylogenetic analysis of PAL3 and its homologs. The phylogenetic analysis was carried out by MEGA X with 1 000 bootstrap replicates and was constructed using the distance method with maximum likelihood.
| Species | Locus ID | Gene name |
|---|---|---|
| Rice | LOC_Os11g12740 | PAL3/SP1 |
| LOC_Os07g01070 | OsPTR1 | |
| LOC_Os12g44100 | OsPTR2 | |
| LOC_Os10g33210 | OsPTR3 | |
| LOC_Os07g41250 | OsPTR4 | |
| LOC_Os04g50940 | OsPTR5 | |
| LOC_Os04g50950 | OsPTR6 | |
| LOC_Os01g04950 | OsPTR7 | |
| LOC_Os03g51050 | OsPTR8 | |
| LOC_Os06g49250 | OsPTR9 | |
| LOC_Os03g13274 | OsNTR1.1 | |
| LOC_Os10g40600 | OsNRT1.1B | |
| LOC_Os01g54515 | OsNPF4.5 | |
| Arabidopsis | At1g12110 | AtNRT1.1 |
| At5g62680 | AtNRT1.10 | |
| At1g52190 | AtNRT1.11 | |
| At1g33440 | AtNRT1.13 | |
| At1g59740 | AtNRT1.14 | |
| At1g72120 | AtNRT1.15 | |
| At1g72125 | AtNRT1.16 | |
| At1g69850 | AtNRT1.2 | |
| At3g21670 | AtNRT1.3 | |
| At2g26690 | AtNRT1.4 | |
| At1g32450 | AtNRT1.5 | |
| At1g27080 | AtNRT1.6 | |
| At1g69870 | AtNRT1.7 | |
| At4g21680 | AtNRT1.8 | |
| At1g18880 | AtNRT1.9 | |
| At3g54140 | AtPTR1 | |
| At2g02040 | AtPTR2/AtNTR1 | |
| At5g46050 | AtPTR3 | |
| At2g02020 | AtPTR4 | |
| At5g01180 | AtPTR5 | |
| At1g62200 | AtPTR6 | |
| At1g27040 | AIT2 | |
| At3g25260 | AIT3 | |
| At3g25280 | AIT4 | |
| At3g47960 | GTR1 | |
| At3g45650 | NAXT1 | |
| At1g68570 | Nitr |
Table 4 Locus ID and gene name of PAL3 and its homologs
| Species | Locus ID | Gene name |
|---|---|---|
| Rice | LOC_Os11g12740 | PAL3/SP1 |
| LOC_Os07g01070 | OsPTR1 | |
| LOC_Os12g44100 | OsPTR2 | |
| LOC_Os10g33210 | OsPTR3 | |
| LOC_Os07g41250 | OsPTR4 | |
| LOC_Os04g50940 | OsPTR5 | |
| LOC_Os04g50950 | OsPTR6 | |
| LOC_Os01g04950 | OsPTR7 | |
| LOC_Os03g51050 | OsPTR8 | |
| LOC_Os06g49250 | OsPTR9 | |
| LOC_Os03g13274 | OsNTR1.1 | |
| LOC_Os10g40600 | OsNRT1.1B | |
| LOC_Os01g54515 | OsNPF4.5 | |
| Arabidopsis | At1g12110 | AtNRT1.1 |
| At5g62680 | AtNRT1.10 | |
| At1g52190 | AtNRT1.11 | |
| At1g33440 | AtNRT1.13 | |
| At1g59740 | AtNRT1.14 | |
| At1g72120 | AtNRT1.15 | |
| At1g72125 | AtNRT1.16 | |
| At1g69850 | AtNRT1.2 | |
| At3g21670 | AtNRT1.3 | |
| At2g26690 | AtNRT1.4 | |
| At1g32450 | AtNRT1.5 | |
| At1g27080 | AtNRT1.6 | |
| At1g69870 | AtNRT1.7 | |
| At4g21680 | AtNRT1.8 | |
| At1g18880 | AtNRT1.9 | |
| At3g54140 | AtPTR1 | |
| At2g02040 | AtPTR2/AtNTR1 | |
| At5g46050 | AtPTR3 | |
| At2g02020 | AtPTR4 | |
| At5g01180 | AtPTR5 | |
| At1g62200 | AtPTR6 | |
| At1g27040 | AIT2 | |
| At3g25260 | AIT3 | |
| At3g25280 | AIT4 | |
| At3g47960 | GTR1 | |
| At3g45650 | NAXT1 | |
| At1g68570 | Nitr |
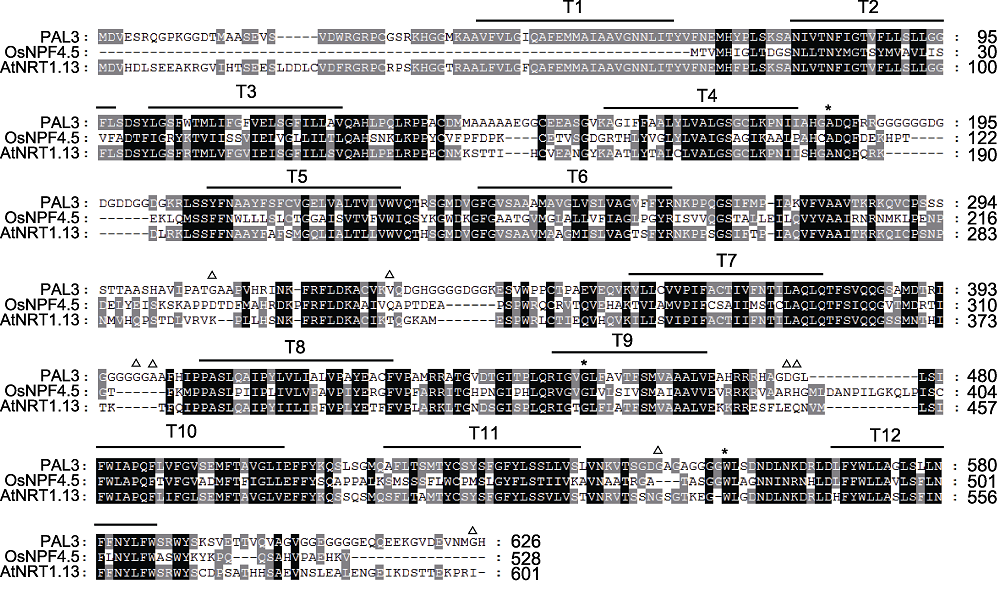
Figure 4 Sequences alignment of PAL3 and its homologs The black background represent that all amino acid sequences are homologous. The gray part represent that some of the amino acid sequences are homologous. The white part represent that the amino acid sequences are completely different. * indicate the mutation sites of the pal3-1, pal3-2 and pal3-4 mutants in turn; △: The variation sites between Hap1 and Hap3.
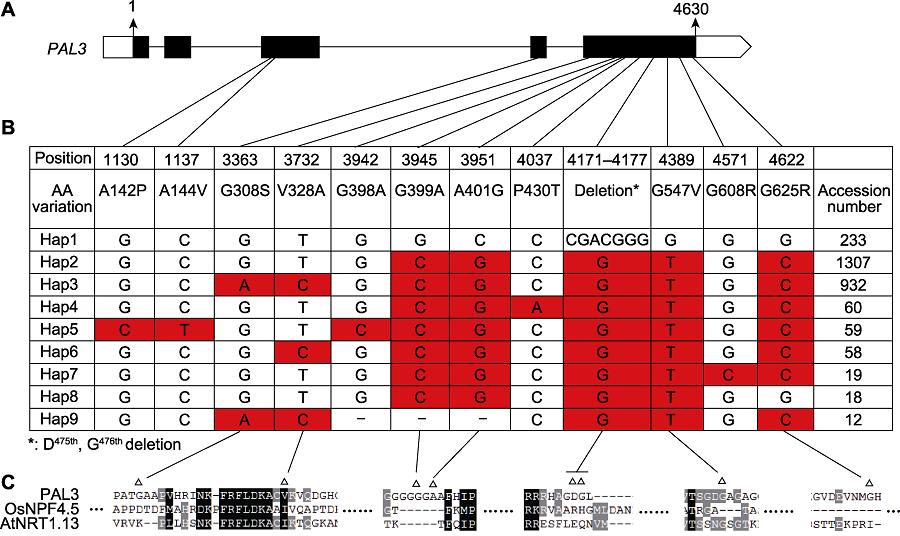
Figure 5 Haplotype analysis of PAL3 (A) Structure of PAL3 gene and the location of haplotypes; (B) Haplotypes of PAL3 in the cultivated rice. Haplotype data were analyzed by Rice Molecular Breeding Knowledgebase (MBKbase) (Peng et al., 2020), including 2 187 cultivated rice varieties. Variations on intron, heterozygous variations and synonymous variations were removed during analysis (-: The deletion of base pair); (C) Sequence alignment around the variation sites between Hap1 and hap3 (Δ: The variation sites between Hap1 and Hap3).
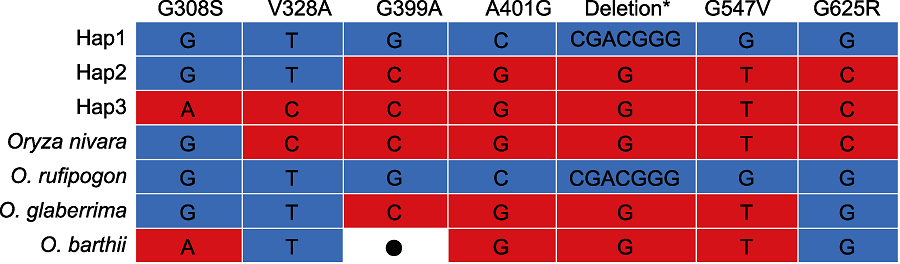
Figure 6 Haplotype of PAL3 in the wild rice Haplotype data of wild rice were from Gramene and Rice Relatives-GD (Mao et al., 2019). *: D475 and G476 deletion; ●: No datum
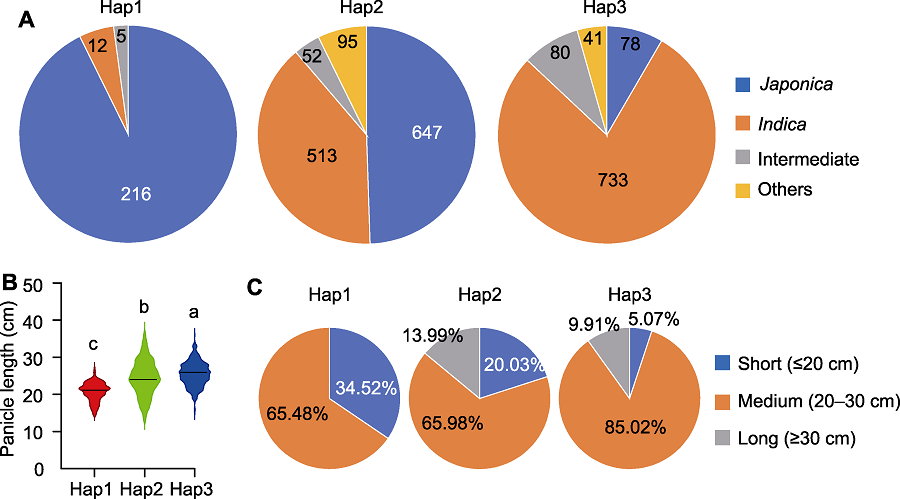
Figure 7 Subspecies composition and panicle phenotype analysis of Hap1, Hap2 and Hap3 of the PAL3 gene (A) Subspecies composition of Hap1, Hap2 and Hap3; (B) Comparison of panicle length among Hap1, Hap2 and Hap3; (C) Statistic of panicle length of Hap1, Hap2 and Hap3. Different lowercase letters indicate significant differences (P<0.05).
| [1] |
淳雁, 李学勇 (2017). 水稻穗型的遗传调控研究进展. 植物学报 52, 19-29.
DOI |
| [2] | 刘丹, 王嘉宇, 刘进, 马殿荣, 赵明辉, 陈温福 (2015). 水稻散状穗突变体sp的遗传分析及基因初定位. 植物学报 50, 198-205. |
| [3] |
刘玉良, 郑术芝 (2017). 水稻产量相关性状驯化研究进展. 植物学报 52, 113-121.
DOI |
| [4] | 田翠 (2010). 复粒稻小穗簇生突变体基因的遗传分析和初步定位. 硕士论文. 重庆: 重庆大学. pp. 19-23. |
| [5] | 吴光南, 张云桥 (1962). 稻穗发育过程及其控制途径的研究. 作物学报 1, 43-52. |
| [6] | 张淑红 (2002). 水稻中控制形态结构建成相关基因的功能研究. 博士论文. 上海: 复旦大学. pp. 70-78. |
| [7] | 郑雷英, 朱旭东, 钱前, 赵忠, 张建军, 胡筱荷, 林鸿宣, 罗达 (2003). 水稻穗部突变体CL的形态和定位分析. 科学通报 48, 264-267. |
| [8] | 周鹏 (2009). 水稻簇生穗突变体Cl-dz的形态特征及遗传定位. 硕士论文. 成都: 四川农业大学. pp. 28-30. |
| [9] |
Chiang CS, Stacey G, Tsay YF (2004). Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J Biol Chem 279, 30150-30157.
DOI URL |
| [10] |
Choi JY, Lye ZN, Groen SC, Dai XG, Rughani P, Zaaijer S, Harrington ED, Juul S, Purugganan MD (2020). Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol 21, 21.
DOI URL |
| [11] | Choi JY, Platts AE, Fuller DQ, Hsing YI, Wing RA, Purugganan MD (2017). The rice paradox: multiple origins but single domestication in Asian rice. Mol Biol Evol 34, 969-979. |
| [12] |
Grillo MA, Li CB, Fowlkes AM, Briggeman TM, Zhou AL, Schemske DW, Sang T (2009). Genetic architecture for the adaptive origin of annual wild rice, Oryza nivala. Evolution 63, 870-883.
DOI URL |
| [13] |
Gross BL, Zhao ZJ (2014). Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci USA 111, 6190-6197.
DOI URL |
| [14] |
Huang XH, Kurata N, Wei XH, Wang ZX, Wang AH, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, Guo YL, Lu YQ, Zhou CC, Fan DL, Weng QJ, Zhu CR, Huang T, Zhang L, Wang YC, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan XP, Xu Q, Dong GJ, Zhan QL, Li CY, Fujiyama A, Toyoda A, Lu TT, Feng Q, Qian Q, Li JY, Han B (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497-501.
DOI URL |
| [15] |
Huang XZ, Qian Q, Liu ZB, Sun HY, He SY, Luo D, Xia GM, Chu CC, Li JY, Fu XD (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41, 494-497.
DOI URL |
| [16] |
Huang Y, Bai XF, Luo MF, Xing YZ (2019). Short Panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice. J Integr Plant Biol 61, 987-999.
DOI |
| [17] |
Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45, 462-465.
DOI URL |
| [18] |
Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003). LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100, 11765-11770.
DOI URL |
| [19] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular Evolutionary Genetics Analysis ac-ross computing platforms. Mol Biol Evol 35, 1547-1549.
DOI URL |
| [20] |
Lee J, Park JJ, Kim SL, Yim J, An G (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65, 487-499.
DOI URL |
| [21] |
Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, Gojon A, Gong JM, Halkier BA, Harris JM, Hedrich R, Limami AM, Rentsch D, Seo M, Tsay YF, Zhang MY, Coruzzi G, Lacombe B (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19, 5-9.
DOI URL |
| [22] |
Li F, Liu WB, Tang JY, Chen JF, Tong HN, Hu B, Li CL, Fang J, Chen MS, Chu CC (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res 20, 838-849.
DOI URL |
| [23] |
Li SB, Qian Q, Fu ZM, Zeng DL, Meng XB, Kyozuka J, Maekawa M, Zhu XD, Zhang J, Li JY, Wang YH (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58, 592-605.
DOI URL |
| [24] |
Liu TM, Li LZ, Zhang YS, Xu CG, Li XH, Xing YZ (2011). Comparison of quantitative trait loci for rice yield, panicle length and spikelet density across three connected populations. J Genet 90, 377-382.
DOI URL |
| [25] | Mao LF, Chen MH, Chu QJ, Jia L, Sultana MH, Wu DY, Kong XD, Qiu J, Ye CY, Zhu QH, Chen X, Fan LJ (2019). RiceRelativesGD: a genomic database of rice relatives for rice research. Database 2019, baz110. |
| [26] |
Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, Boyko AR, Purugganan MD (2011). Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA 108, 8351-8356.
DOI URL |
| [27] |
Nakagawa M, Shimamoto K, Kyozuka J (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29, 743-750.
PMID |
| [28] | Peng H, Wang K, Chen Z, Cao YH, Gao Q, Li Y, Li XX, Lu HW, Du HL, Lu M, Yang X, Liang CZ (2020). MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res 48, D1085-D1092. |
| [29] |
Piao RH, Jiang WZ, Ham TH, Choi MS, Qiao YL, Chu SH, Park JH, Woo MO, Jin ZX, An G, Lee J, Koh HJ (2009). Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor Appl Genet 119, 1497-1506.
DOI URL |
| [30] |
Qiao YL, Piao RH, Shi JX, Lee SI, Jiang WZ, Kim BK, Lee J, Han LZ, Ma WB, Koh HJ (2011). Fine mapping and candidate gene analysis of Dense and Erect Panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor Appl Genet 122, 1439-1449.
DOI URL |
| [31] |
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang XH, Yoshida H, Kyozuka J, Chen F, Sato Y (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276-3287.
DOI URL |
| [32] |
Vaughan DA, Morishima H, Kadowaki K (2003). Diversity in the Oryza genus. Curr Opin Plant Biol 6, 139-146.
PMID |
| [33] |
Wu YZ, Fu YC, Zhao SS, Gu P, Zhu ZF, Sun CQ, Tan LB (2016). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J 14, 377-386.
DOI URL |
| [34] |
Yan CJ, Zhou JH, Yan S, Chen F, Yeboah M, Tang SZ, Liang GH, Gu MH (2007). Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor Appl Genet 115, 1093-1100.
DOI URL |
| [35] |
Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen RH, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, Nagamura Y, Ushijima T, Kumamaru T, Iida S, Maekawa M, Kyozuka J (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110, 767-772.
DOI URL |
| [36] |
Yu HY, Murchie EH, González-carranza ZH, Pyke KA, Roberts JA (2015). Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development. J Exp Bot 66, 1543-1552.
DOI URL |
| [37] |
Zhou Y, Zhu JY, Li ZY, Yi CD, Liu J, Zhang HG, Tang SZ, Gu MH, Liang GH (2009). Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183, 315-324.
DOI URL |
| [38] |
Zhu ZF, Tan LB, Fu YC, Liu FX, Cai HW, Xie DX, Wu F, Wu JZ, Matsumoto T, Sun CQ (2013). Genetic control of inflorescence architecture during rice domestication. Nat Commun 4, 2200.
DOI URL |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [3] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [4] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [5] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [6] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [7] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [8] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [9] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [10] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [11] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [12] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [13] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [14] | Yuqiang Liu, Jianmin Wan. The Host Controls the Protein Level of Insect Effectors to Balance Immunity and Growth [J]. Chinese Bulletin of Botany, 2023, 58(3): 353-355. |
| [15] | Jingjing Zhao, Haibin Jia, Tien Ming Lee. Market status and the sustainable utilization strategy of wild earthworm (earth dragon) for medicinal use [J]. Biodiv Sci, 2023, 31(3): 22478-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||