

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (3): 351-368.DOI: 10.11983/CBB19135 cstr: 32102.14.CBB19135
• SPECIAL TOPICS • Previous Articles Next Articles
Yu Zhang1,2,3,Mingjie Zhao1,2,3,Wei Zhang1,2,3,*( )
)
Received:2019-07-15
Accepted:2020-03-23
Online:2020-05-01
Published:2020-07-06
Contact:
Wei Zhang
Yu Zhang, Mingjie Zhao, Wei Zhang. Transcriptional Regulatory Network of Secondary Cell Wall Biosynthesis in Plants[J]. Chinese Bulletin of Botany, 2020, 55(3): 351-368.
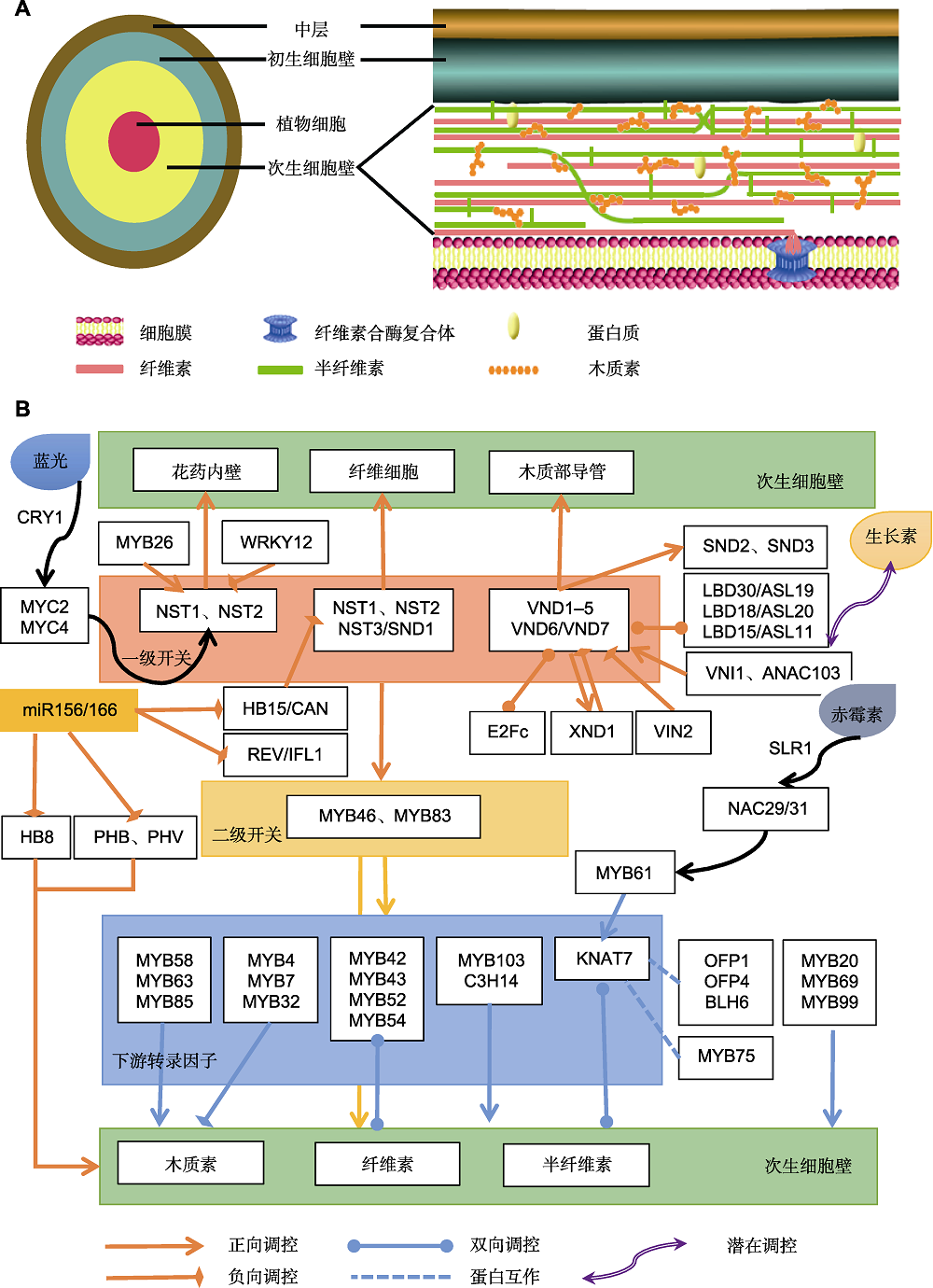
Figure 1 Structure of secondary cell wall and its biosynthetic transcriptional regulatory network in Arabidopsis thaliana (A) Structural sketch of the secondary cell wall; (B) Transcriptional regulatory network of the secondary cell wall biosynthesis
| [1] | 李慧, 刘凤栾, 郗琳, 高彬, 严珊, 王玲, 马男, 赵梁军, 杨春起 (2012). 月季皮刺的组织结构与化学组成. 园艺学报 39, 1321-1329. |
| [2] |
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841-857.
URL PMID |
| [3] | Asano G, Kubo R, Tanimoto S (2008). Growth, structure and lignin localization in rose prickle. Bull Fac Agric Saga Univ (93), 117-125. |
| [4] |
Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001). The Arabidopsis ATHB- 8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126, 643-655.
URL PMID |
| [5] |
Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B (2012). SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22, 640-654.
DOI URL PMID |
| [6] |
Bhargava A, Ahad A, Wang SC, Mansfield SD, Haughn GW, Douglas CJ, Ellis BE (2013). The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 237, 1199-1211.
DOI URL PMID |
| [7] |
Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE (2010). MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 154, 1428-1438.
DOI URL PMID |
| [8] |
Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency M, Pavy N, Grima-Pettenati J, Séguin A, MacKay J (2008). Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot 59, 3925-3939.
DOI URL PMID |
| [9] |
Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383-2393.
DOI URL PMID |
| [10] |
Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281-2295.
DOI URL PMID |
| [11] |
Cassan-Wang H, Goué N, Saidi MN, Legay S, Sivadon P, Goffner D, Grima-Pettenati J (2013). Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front Plant Sci 4, 189.
DOI URL PMID |
| [12] |
Chai GH, Qi G, Cao YP, Wang ZG, Yu L, Tang XF, Yu YC, Wang D, Kong YZ, Zhou GK (2014). Poplar PdC3H17 and PdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol 203, 520-534.
DOI URL PMID |
| [13] |
Christianson JA, Dennis ES, Llewellyn DJ, Wilson IW (2010). ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signal Behav 5, 428-432.
DOI URL PMID |
| [14] |
Cosgrove DJ, Jarvis MC (2012). Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3, 204.
DOI URL PMID |
| [15] | Dawson J, Sözen E, Vizir I, Waeyenberge SV, Wilson ZA, Mulligan BJ (1999). Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol 144, 213-222. |
| [16] |
Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y, Sassa N, Suzuki S, Yazaki J, Kikuchi S, Fukuda H (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA 99, 15794-15799.
URL PMID |
| [17] |
Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002). Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43, 1407-1420.
DOI URL PMID |
| [18] |
Du Q, Avci U, Li SB, Gallego-Giraldo L, Pattathil S, Qi LY, Hahn MG, Wang HZ (2015). Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis. Plant J 83, 388-400.
DOI URL PMID |
| [19] |
Du Q, Wang HZ (2015). The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal Behav 10, e1078955.
DOI URL PMID |
| [20] |
Endo H, Yamaguchi M, Tamura T, Nakano Y, Nishikubo N, Yoneda A, Kato K, Kubo M, Kajita S, Katayama Y, Ohtani M, Demura T (2015). Multiple classes of transcription factors regulate the expression of VASCULAR- RELATED NAC-DOMAIN 7, a master switch of xylem vessel differentiation. Plant Cell Physiol 56, 242-254.
DOI URL PMID |
| [21] |
Ernst HA, Olsen AN, Skriver K, Larsen S, Lo Leggio L (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5, 297-303.
DOI URL PMID |
| [22] |
Fukuda H (2004). Signals that control plant vascular cell differentiation. Nat Rev Mol Cell Biol 5, 379-391.
DOI URL PMID |
| [23] |
Gallego-Giraldo L, Shadle G, Shen H, Barros-Rios J, Corrales SF, Wang HZ, Dixon RA (2016). Combining enhanced biomass density with reduced lignin level for improved forage quality. Plant Biotechnol J 14, 895-904.
DOI URL PMID |
| [24] |
Goubet F, Barton CJ, Mortimer JC, Yu XL, Zhang ZN, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60, 527-538.
DOI URL PMID |
| [25] |
Grant EH, Fujino T, Beers EP, Brunner AM (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337-352.
DOI URL PMID |
| [26] | Guo Y, Cai Z, Gan S (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27, 521-549. |
| [27] |
Handakumbura PP, Hazen SP (2012). Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front Plant Sci 3, 74.
DOI URL PMID |
| [28] |
Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033-1043.
DOI URL PMID |
| [29] |
He JB, Zhao XH, Du PZ, Zeng W, Beahan CT, Wang YQ, Li HL, Bacic A, Wu AM (2018). KNAT7 positively regulates xylan biosynthesis by directly activating IRX9 expression in Arabidopsis. J Integr Plant Biol 60, 514-528.
DOI URL |
| [30] |
Huang DB, Wang SG, Zhang BC, Shangguan KK, Shi YY, Zhang DM, Liu XL, Wu K, Xu ZP, Fu XD, Zhou YH (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681-1696.
DOI URL PMID |
| [31] |
Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol 11, 173.
DOI URL PMID |
| [32] |
Itoh JI, Hibara KI, Sato Y, Nagato Y (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147, 1960-1975.
DOI URL PMID |
| [33] |
Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000). Transcriptional repression by AtMYB4 controls production of UV- protecting sunscreens in Arabidopsis. EMBO J 19, 6150-6161.
DOI URL PMID |
| [34] |
Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Kim VN, Chua NH, Park CM (2005). MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42, 84-94.
DOI URL PMID |
| [35] |
Kim WC, Kim JY, Ko JH, Kang H, Han KH (2014a). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol Biol 85, 589-599.
DOI URL PMID |
| [36] |
Kim WC, Kim JY, Ko JH, Kang H, Kim J, Han KH (2014b). AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mRNAs in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. Plant J 80, 772-784.
DOI URL PMID |
| [37] |
Kim WC, Kim JY, Ko JH, Kim J, Han KH (2013a). Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J Plant Physiol 170, 1374-1378.
DOI URL PMID |
| [38] |
Kim WC, Ko JH, Han KH (2012). Identification of a cis- acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol Biol 78, 489-501.
DOI URL PMID |
| [39] |
Kim WC, Ko JH, Kim JY, Kim J, Bae HJ, Han KH (2013b). MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J 73, 26-36.
DOI URL PMID |
| [40] |
Kim WC, Reca IB, Kim Y, Park S, Thomashow MF, Keegstra K, Han KH (2014c). Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol Biol 84, 577-587.
DOI URL PMID |
| [41] |
Ko JH, Jeon HW, Kim WC, Kim JY, Han KH (2014). The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot 114, 1099-1107.
DOI URL PMID |
| [42] |
Ko JH, Kim WC, Han KH (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60, 649-665.
DOI URL PMID |
| [43] |
Ko JH, Kim WC, Kim JY, Ahn SJ, Han KH (2012). MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol Plant 5, 961-963.
DOI URL PMID |
| [44] |
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH (2007). ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50, 1035-1048.
DOI URL PMID |
| [45] |
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19, 1855-1860.
URL PMID |
| [46] |
Li EY, Bhargava A, Qiang WY, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ (2012). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194, 102-115.
DOI URL PMID |
| [47] |
Li EY, Wang SC, Liu YY, Chen JG, Douglas CJ (2011). OVATE FAMILY PROTEIN 4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67, 328-341.
DOI URL PMID |
| [48] |
Li W, Huang GQ, Zhou W, Xia XC, Li DD, Li XB (2014). A cotton (Gossypium hirsutum) gene encoding a NAC transcription factor is involved in negative regulation of plant xylem development. Plant Physiol Biochem 83, 134-141.
DOI URL PMID |
| [49] |
Liepman AH, Wilkerson CG, Keegstra K (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102, 2221-2226.
DOI URL PMID |
| [50] |
Liu JY, Osbourn A, Ma PD (2015a). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8, 689-708.
DOI URL PMID |
| [51] |
Liu YY, Douglas CJ (2015). A role for OVATE FAMILY PROTEIN 1 (OFP1) and OFP4 in a BLH6-KNAT7 multi- protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal Behav 10, e1033126.
DOI URL PMID |
| [52] |
Liu YY, You SJ, Taylor-Teeples M, Li WL, Schuetz M, Brady SM, Douglas CJ (2015b). BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 26, 4843-4861.
DOI URL PMID |
| [53] |
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang GL, Zamore PD, Barton MK, Bartel DP (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J 23, 3356-3364.
DOI URL PMID |
| [54] |
McCarthy RL, Zhong RQ, Fowler S, Lyskowski D, Piyasena H, Carleton K, Spicer C, Ye ZH (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol 51, 1084-1090.
DOI URL PMID |
| [55] |
McCarthy RL, Zhong RQ, Ye ZH (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50, 1950-1964.
DOI URL PMID |
| [56] |
McCarthy RL, Zhong RQ, Ye ZH (2011). Secondary wall NAC binding element (SNBE), a key cis-acting element required for target gene activation by secondary wall NAC master switches. Plant Signal Behav 6, 1282-1285.
DOI URL PMID |
| [57] |
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270-280.
DOI URL PMID |
| [58] |
Mitsuda N, Ohme-Takagi M (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56, 768-778.
DOI URL PMID |
| [59] |
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993-3006.
DOI URL PMID |
| [60] |
Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M (2005). Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96, 673-686.
DOI URL PMID |
| [61] | Nakano Y, Nishikubo N, Goué N, Ohtani M, Yamaguchi M, Katayama Y, Demura T (2010). MYB transcription factors orchestrating the developmental program of xylem vessels in Arabidopsis roots. Plant Biotechnol 27, 267-272. |
| [62] |
Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6, 288.
DOI URL PMID |
| [63] |
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012). NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta (BBA)- Gene Regul Mech 1819, 97-103.
DOI URL PMID |
| [64] |
Ohashi-Ito K, Fukuda H (2003). HD-ZIP III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44, 1350-1358.
DOI URL PMID |
| [65] |
Ohashi-Ito K, Kubo M, Demura T, Fukuda H (2005). Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 46, 1646-1656.
DOI URL PMID |
| [66] |
Ohashi-Ito K, Oda Y, Fukuda H (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461-3473.
DOI URL PMID |
| [67] |
Öhman D, Demedts B, Kumar M, Gerber L, Gorzsás A, Goeminne G, Hedenström M, Ellis B, Boerjan W, Sundberg B (2013). MYB103 is required for FERULATE- 5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73, 63-76.
DOI URL |
| [68] |
Ohtani M, Nishikubo N, Xu B, Yamaguchi M, Mitsuda N, Goué N, Shi FS, Ohme-Takagi M, Demura T (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J 67, 499-512.
DOI URL |
| [69] |
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10, 79-87.
DOI URL PMID |
| [70] |
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10, 239-247.
DOI URL PMID |
| [71] |
Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40, 979-995.
DOI URL PMID |
| [72] |
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17, 369-381.
DOI URL PMID |
| [73] |
Pyo H, Demura T, Fukuda H (2007). TERE; a novel cis- element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J 51, 955-965.
DOI URL PMID |
| [74] |
Rao X, Dixon RA (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front Plant Sci 9, 399.
DOI URL PMID |
| [75] |
Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang KY, Li EY, Douglas CJ, Western TL, Mansfield SD, Campbell MM (2012). AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol 195, 774-786.
DOI URL PMID |
| [76] |
Sakamoto S, Mitsuda N (2014). Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol 56, 299-310.
DOI URL PMID |
| [77] |
Sakamoto S, Takata N, Oshima Y, Yoshida K, Taniguchi T, Mitsuda N (2016). Wood reinforcement of poplar by rice NAC transcription factor. Sci Rep 6, 19925.
DOI URL PMID |
| [78] |
Shao HB, Wang HY, Tang XL (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6, 902.
DOI URL PMID |
| [79] |
Souer E, Van Houwelingen A, Kloos D, Mol J, Koes R (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159-170.
DOI URL PMID |
| [80] |
Soyano T, Thitamadee S, Machida Y, Chua NH (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359-3373.
DOI URL PMID |
| [81] |
Steiner-Lange S, Unte US, Eckstein L, Yang CY, Wilson ZA, Schmelzer E, Dekker K, Saedler H (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34, 519-528.
DOI URL PMID |
| [82] |
Takata N, Awano T, Nakata MT, Sano Y, Sakamoto S, Mitsuda N, Taniguchi T (2019). Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol 39, 514-525.
DOI URL PMID |
| [83] |
Takata N, Sakamoto S, Mitsuda N, Taniguchi T (2017). The Arabidopsis NST3/SND1 promoter is active in secondary woody tissue in poplar. J Wood Sci 63, 396-400.
DOI URL |
| [84] |
Tan TT, Endo H, Sano R, Kurata T, Yamaguchi M, Ohtani M, Demura T (2018). Transcription factors VND1-VND3 contribute to cotyledon xylem vessel formation. Plant Physiol 176, 773-789.
DOI URL PMID |
| [85] |
Tang N, Shahzad Z, Lonjon F, Loudet O, Vailleau F, Maurel C (2018). Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nat Commun 9, 3884.
DOI URL PMID |
| [86] |
Tang XF, Zhuang YM, Qi G, Wang D, Liu HH, Wang KR, Chai GH, Zhou GK (2015). Poplar PdMYB221 is involved in the direct and indirect regulation of secondary wall biosynthesis during wood formation. Sci Rep 5, 12240.
DOI URL PMID |
| [87] |
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100, 1450-1455.
DOI URL PMID |
| [88] |
Taylor NG, Laurie S, Turner SR (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529-2539.
DOI URL PMID |
| [89] |
Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769-779.
DOI URL PMID |
| [90] |
Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda-Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571-575.
DOI URL PMID |
| [91] |
Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32-39.
DOI URL PMID |
| [92] |
Wang HZ, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 107, 22338-22343.
DOI URL PMID |
| [93] |
Wang HZ, Dixon RA (2012). On-off switches for secondary cell wall biosynthesis. Mol Plant 5, 297-303.
DOI URL PMID |
| [94] |
Wang LJ, Lu WX, Ran LY, Dou LW, Yao S, Hu J, Fan D, Li CF, Luo KM (2019). R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J 99, 733-751.
DOI URL PMID |
| [95] |
Welner DH, Lindemose S, Grossmann JG, Møllegaard NE, Olsen AN, Helgstrand C, Skriver K, Leggio LL (2012). DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem J 444, 395-404.
DOI URL PMID |
| [96] |
Willemsen V, Bauch M, Bennett T, Campilho A, Wolkenfelt H, Xu J, Haseloff J, Scheres B (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell 15, 913-922.
DOI URL PMID |
| [97] |
Williams L, Grigg SP, Xie MT, Christensen S, Fletcher JC (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132, 3657-3668.
DOI URL PMID |
| [98] |
Williamson RE, Burn JE, Hocart CH (2002). Towards the mechanism of cellulose synthesis. Trends Plant Sci 7, 461-467.
DOI URL PMID |
| [99] |
Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, Murata T, Kurata T, Yoneda A, Kato K, Hasebe M, Demura T (2014). Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505-1508.
DOI URL PMID |
| [100] |
Yamaguchi M, Demura T (2010). Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol 27, 237-242.
DOI URL |
| [101] |
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008). VASCULAR-RELATED NAC-DOMAIN 7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55, 652-664.
DOI URL PMID |
| [102] |
Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T (2011). VASCULAR-RELATED NAC- DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66, 579-590.
DOI URL PMID |
| [103] |
Yamaguchi M, Nagahage ISP, Ohtani M, Ishikawa T, Uchimiya H, Kawai-Yamada M, Demura T (2015). Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnol 32, 119-123.
DOI URL |
| [104] |
Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme- Takagi M, Fukuda H, Demura T (2010c). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249-1263.
DOI URL PMID |
| [105] |
Yan L, Xu CH, Kang YL, Gu TW, Wang DX, Zhao SY, Xia GM (2013). The heterologous expression in Arabidopsis thaliana of sorghum transcription factor SbbHLH1 downregulates lignin synthesis. J Exp Bot 64, 3021-3032.
DOI URL PMID |
| [106] |
Yang CY, Xu ZY, Song J, Conner K, Barrena GV, Wilson ZA (2007). Arabidopsis MYB26/MALESTERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534-548.
DOI URL PMID |
| [107] |
Yang JH, Wang HZ (2016). Molecular mechanisms for vascular development and secondary cell wall formation. Front Plant Sci 7, 356.
DOI URL PMID |
| [108] |
Yang SD, Seo PJ, Yoon HK, Park CM (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23, 2155-2168.
DOI URL PMID |
| [109] |
Ye YF, Wu K, Chen JF, Liu Q, Wu YJ, Liu BM, Fu XD (2018). OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 11, 36.
DOI URL PMID |
| [110] |
Zhang DM, Xu ZP, Cao SX, Chen KL, Li SC, Liu XL, Gao CX, Zhang BC, Zhou YH (2018a). An uncanonical CCCH-tandem zinc-finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol Plant 11, 163-174.
URL PMID |
| [111] |
Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, Hu S, Li XB (2018b). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol 217, 625-640.
DOI URL PMID |
| [112] |
Zhang J, Xie M, Tuskan GA, Muchero W, Chen JG (2018c). Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Front Plant Sci 9, 1535.
DOI URL PMID |
| [113] |
Zhang Q, Xie Z, Zhang R, Xu P, Liu HT, Yang HQ, Doblin MS, Bacic A, Li LG (2018d). Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-directed transcriptional network in Arabidopsis. Plant Cell 30, 2512-2528.
DOI URL PMID |
| [114] |
Zhao CS, Avci U, Grant EH, Haigler CH, Beers EP (2008). XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J 53, 425-436.
DOI URL PMID |
| [115] |
Zhao CS, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol 138, 803-818.
URL PMID |
| [116] |
Zhao CS, Lasses T, Bako L, Kong DY, Zhao BY, Chanda B, Bombarely A, Cruz-Ramírez A, Scheres B, Brunner AM, Beers EP (2017). XYLEM NAC DOMAIN 1, an angiosperm NAC transcription factor, inhibits xylem differentiation through conserved motifs that interact with RETINOBLASTOMA-RELATED. New Phytol 216, 76-89.
DOI URL PMID |
| [117] |
Zhao KM, Bartley LE (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol 14, 135.
DOI URL PMID |
| [118] |
Zhao YQ, Song XQ, Zhou HJ, Wei KL, Jiang C, Wang JN, Cao Y, Tang F, Zhao ST, Lu MZ (2020). KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol 225, 1531-1544.
URL PMID |
| [119] |
Zhong RQ, Demura T, Ye ZH (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158-3170.
DOI URL PMID |
| [120] |
Zhong RQ, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 52, 1856-1871.
DOI URL PMID |
| [121] |
Zhong RQ, Lee C, Ye ZH (2010a). Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 152, 1044-1055.
DOI URL PMID |
| [122] |
Zhong RQ, Lee C, Ye ZH (2010b). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3, 1087-1103.
DOI URL PMID |
| [123] |
Zhong RQ, Lee C, Ye ZH (2010c). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15, 625-632.
DOI URL PMID |
| [124] |
Zhong RQ, Lee C, Zhou JL, McCarthy RL, Ye ZH (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763-2782.
DOI URL PMID |
| [125] |
Zhong RQ, McCarthy RL, Haghighat M, Ye ZH (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8, e69219.
DOI URL PMID |
| [126] |
Zhong RQ, Richardson EA, Ye ZH (2007a). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225, 1603-1611.
DOI URL PMID |
| [127] |
Zhong RQ, Richardson EA, Ye ZH (2007b). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776-2792.
DOI URL PMID |
| [128] |
Zhong RQ, Ye ZH (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53, 368-380.
DOI URL PMID |
| [129] |
Zhong RQ, Ye ZH (2014). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56, 195-214.
URL PMID |
| [130] |
Zhong RQ, Ye ZH (2015). The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal Behav 10, e989746.
DOI URL PMID |
| [131] |
Zhong RQ, Yuan YX, Spiekerman JJ, Guley JT, Egbosiuba JC, Ye ZH (2015). Functional characterization of NAC and MYB transcription factors involved in regulation of biomass production in Switchgrass (Panicum virgatum). PLoS One 10, e0134611.
DOI URL PMID |
| [132] |
Zhou JL, Lee C, Zhong RQ, Ye ZH (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248-266.
DOI URL PMID |
| [133] |
Zhou JL, Zhong RQ, Ye ZH (2014). Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9, e105726.
DOI URL PMID |
| [1] | Zhou Jing, Gao Fei. Advances in Iron Deficiency-induced Coumarin Biosynthesis and Their Functions in Iron Absorption in Plants [J]. Chinese Bulletin of Botany, 2025, 60(3): 460-471. |
| [2] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [3] | Haitao Hu, Yue Wu, Ling Yang. Research Progress on the NAD(P)+ Biosynthesis and Function in Plants [J]. Chinese Bulletin of Botany, 2025, 60(1): 114-131. |
| [4] | Xuelan Fan, Yanjiao Luo, Chaoqun Xu, Baolin Guo. Research Progress on Genes Related to Flavonoids Biosynthesis in Herba Epimedii [J]. Chinese Bulletin of Botany, 2024, 59(5): 834-846. |
| [5] | Jing Xia, Yuchun Rao, Danyun Cao, Yi Wang, Linxin Liu, Yating Xu, Wangshu Mou, Dawei Xue. Research Progress on the Regulatory Mechanisms of OsACS and OsACO in Rice Ethylene Biosynthesis [J]. Chinese Bulletin of Botany, 2024, 59(2): 291-301. |
| [6] | Xiaoxiao Liu, Di Gong, Tianpeng Gao, Lina Yin, Shiwen Wang. The Major Membrane Lipids in Plant Thylakoids and Their Biosynthesis [J]. Chinese Bulletin of Botany, 2024, 59(1): 144-155. |
| [7] | Haitao Hu, Longbiao Guo. Progress in the Research on Riboflavin Biosynthesis and Function in Plants [J]. Chinese Bulletin of Botany, 2023, 58(4): 638-655. |
| [8] | Yanjun Guo, Feng Chen, Jingwen Luo, Wei Zeng, Wenliang Xu. The Biosynthesis of Plant Cell Wall Xylan and Its Application [J]. Chinese Bulletin of Botany, 2023, 58(2): 316-334. |
| [9] | Yu Miao, Ruan Chengjiang, Ding Jian, Li Jingbin, Lu Shunguang, Wen Xiufeng. Hrh-miRn458 Regulates Oil Biosynthesis of Sea Buckthorn via Targeting Transcription Factor WRI1 [J]. Chinese Bulletin of Botany, 2022, 57(5): 635-648. |
| [10] | Jingwen Wang, Xingjun Wang, Changle Ma, Pengcheng Li. A Review on the Mechanism of Ribosome Stress Response in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 80-89. |
| [11] | Deshuai Liu, Lei Yao, Weirong Xu, Mei Feng, Wenkong Yao. Research Progress of Melatonin in Plant Stress Resistance [J]. Chinese Bulletin of Botany, 2022, 57(1): 111-126. |
| [12] | Kaicheng Kang, Xiqiang Niu, Xianzhong Huang, Nengbing Hu, Yihu Sui, Kaijing Zhang, Hao Ai. Genome-wide Identification and Comparative Evolutionary Analysis of the R2R3-MYB Transcription Factor Gene Family in Pepper [J]. Chinese Bulletin of Botany, 2021, 56(3): 315-329. |
| [13] | Peipei Liu, Geng Zhang, Xiaojuan Li. Biosynthesis and Function of Plant Pectin [J]. Chinese Bulletin of Botany, 2021, 56(2): 191-200. |
| [14] | Yanmei Dong, Wenying Zhang, Zhengyi Ling, Jingrui Li, Hongtong Bai, Hui Li, Lei Shi. Advances in Transcription Factors Regulating Plant Terpenoids Biosynthesis [J]. Chinese Bulletin of Botany, 2020, 55(3): 340-350. |
| [15] | Wei Chen,Xiaoxian Zeng,Chuping Xie,Chang’en Tian,Yuping Zhou. The Dynamic Regulation Mechanism of the Endo-genous ABA in Plant [J]. Chinese Bulletin of Botany, 2019, 54(6): 677-687. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||