

植物学报 ›› 2025, Vol. 60 ›› Issue (2): 256-270.DOI: 10.11983/CBB24067 cstr: 32102.14.CBB24067
收稿日期:2024-05-05
接受日期:2024-10-14
出版日期:2025-03-10
发布日期:2024-10-21
通讯作者:
包劲松
基金资助:
Xinyu Li1, Yue Gu1, Feifei Xu1, Jinsong Bao1,2,*( )
)
Received:2024-05-05
Accepted:2024-10-14
Online:2025-03-10
Published:2024-10-21
Contact:
Jinsong Bao
摘要: 蛋白质翻译后修饰(PTMs)是调控蛋白质生物学功能的重要机制, 在水稻(Oryza sativa)种子发育和胚乳淀粉生物合成中起重要作用。随着蛋白质组学的发展, 已在水稻胚乳中鉴定到大量淀粉合成相关酶(SSREs)发生蛋白质PTMs。该文总结了水稻胚乳SSREs的磷酸化、赖氨酸乙酰化、琥珀酰化、2-羟基异丁酰化、丙二酰化及泛素化6种PTMs的蛋白质组学分析、修饰位点和途径及生物学功能。其中, 蛋白质磷酸化修饰的研究最多, 其在调节植物生长发育和淀粉合成代谢过程中起关键作用。此外, 还讨论了PTMs对籽粒灌浆、稻米淀粉品质及外观的潜在作用。该文综述了PTMs在水稻胚乳淀粉合成相关蛋白中的调控机制, 为培育高产优质水稻品种提供了有价值的参考。
李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展. 植物学报, 2025, 60(2): 256-270.
Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm. Chinese Bulletin of Botany, 2025, 60(2): 256-270.
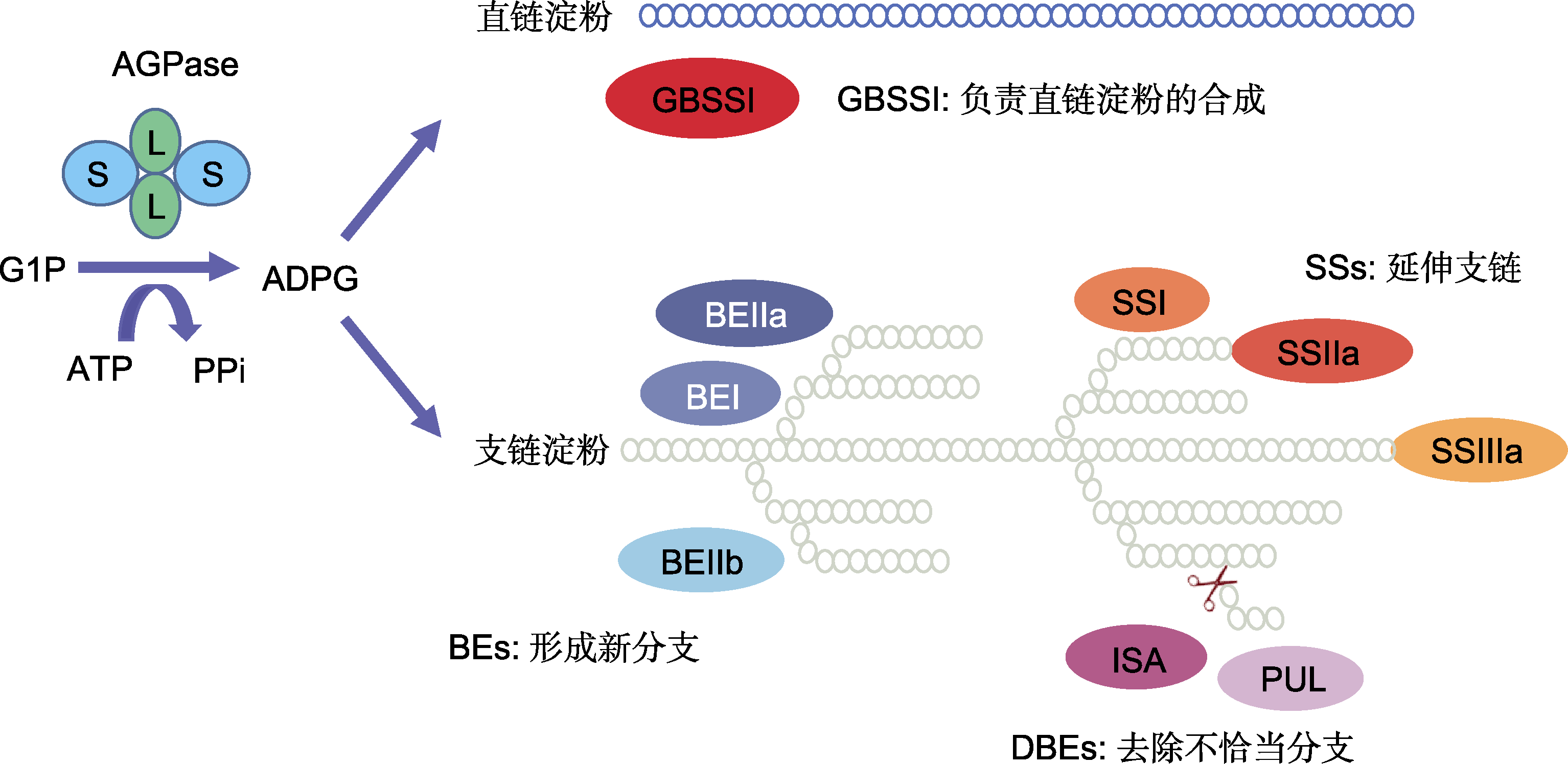
图1 水稻胚乳淀粉生物合成相关酶的功能 G1P: 葡萄糖-1-磷酸; ADPG: 腺苷二磷酸葡萄糖; AGPase: ADP-葡萄糖焦磷酸化酶; GBSSI: 颗粒结合淀粉合酶I; SSs: 可溶性淀粉合酶; BEs: 淀粉分支酶; DBEs: 淀粉去分支酶; ISA: 异淀粉酶; PUL: 普鲁兰酶
Figure 1 The enzyme functions involved in starch biosynthesis in rice endosperm G1P: Glucose-1-phosphate; ADPG: Adenosine diphosphate glucose; AGPase: ADP-glucose pyrophosphorylase; GBSSI: Granule-bound starch synthase I; SSs: Soluble starch synthases; BEs: Branching enzymes; DBEs: Debranching enzymes; ISA: Isoamylase; PUL: Pullulanase
| 修饰类型 | 蛋白数量 | 位点数量 |
|---|---|---|
| 磷酸化 | 7796 | 23513 |
| 赖氨酸2-羟基异丁酰化 | 4719 | 19831 |
| 赖氨酸乙酰化 | 2607 | 4798 |
| 赖氨酸琥珀酰化 | 859 | 2381 |
| 赖氨酸丙二酰化 | 483 | 898 |
| 赖氨酸类泛素化 | 403 | 978 |
| N-糖基化 | 202 | 248 |
| 羰基化 | 181 | 414 |
表1 水稻蛋白质翻译后修饰(PTMs)相关蛋白及位点数量
Table 1 The number of proteins and sites associated with post-translational modifications (PTMs) of rice
| 修饰类型 | 蛋白数量 | 位点数量 |
|---|---|---|
| 磷酸化 | 7796 | 23513 |
| 赖氨酸2-羟基异丁酰化 | 4719 | 19831 |
| 赖氨酸乙酰化 | 2607 | 4798 |
| 赖氨酸琥珀酰化 | 859 | 2381 |
| 赖氨酸丙二酰化 | 483 | 898 |
| 赖氨酸类泛素化 | 403 | 978 |
| N-糖基化 | 202 | 248 |
| 羰基化 | 181 | 414 |
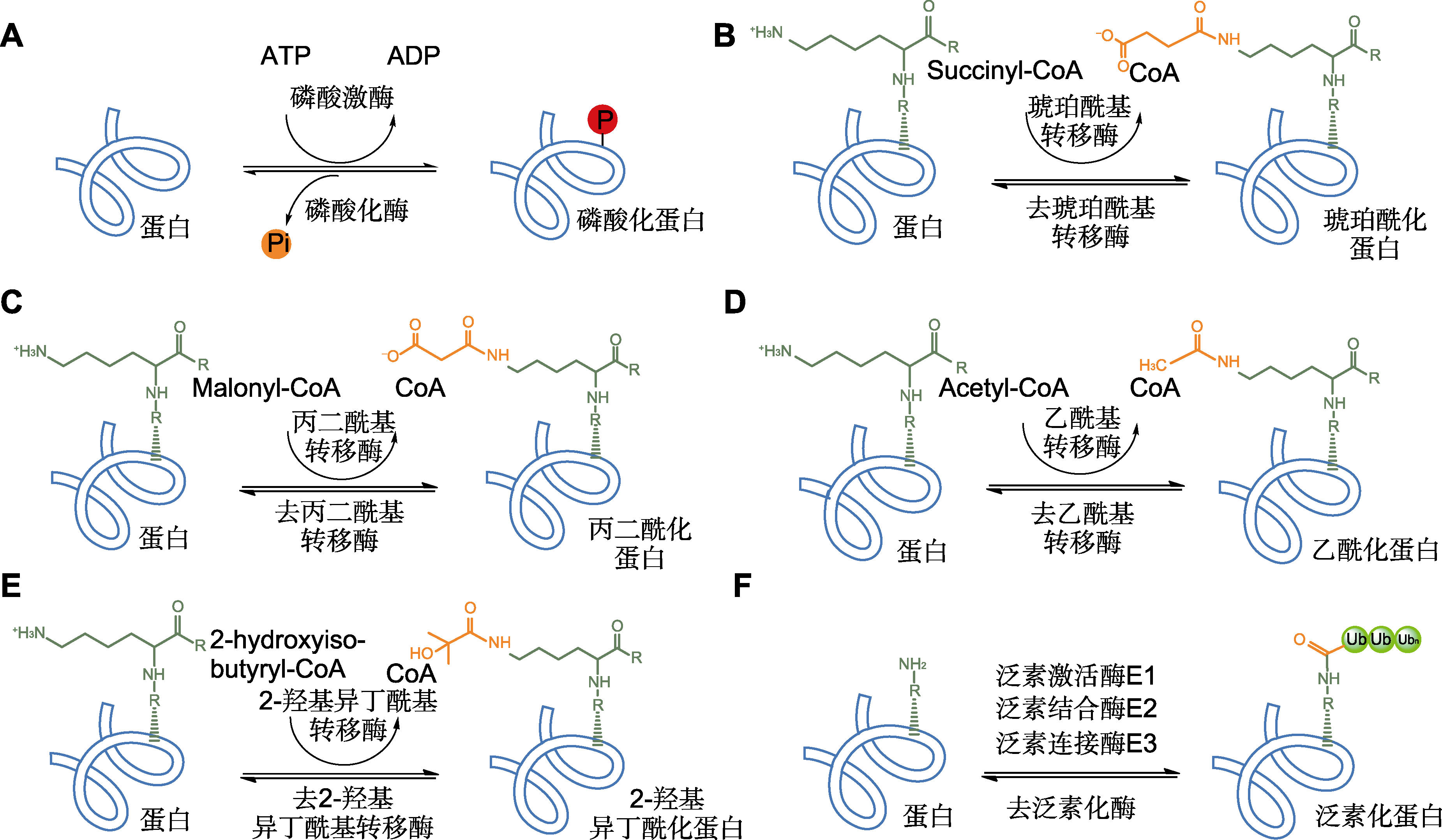
图2 从水稻种子中鉴定到的6种常见蛋白质翻译后修饰(PTMs) (A) 磷酸化修饰; (B) 琥珀酰化修饰; (C) 丙二酰化修饰; (D) 乙酰化修饰; (E) 2-羟基异丁酰化修饰; (F) 泛素化修饰
Figure 2 Six common types of post-translational modifications (PTMs) identified from rice seeds (A) Phosphorylation; (B) Succinylation; (C) Malonylation; (D) Acetylation; (E) 2-hydroxyisobutyrylation; (F) Ubiquitination
| 蛋白质磷酸化类型 | 蛋白质磷酸化位点 | 化学键 |
|---|---|---|
| O-磷酸化 | 苏氨酸(Thr) | P-O |
| 丝氨酸(Ser) | ||
| 酪氨酸(Tyr) | ||
| N-磷酸化 | 组氨酸(His) | P-N |
| 精氨酸(Arg) | ||
| 赖氨酸(Lys) | ||
| S-磷酸化 | 谷氨酸(Glu) | P-O |
| 天冬氨酸(Asp) | ||
| 酰基磷酸化 | 半胱氨酸(Cys) | P-S |
表2 蛋白质磷酸化类型及位点
Table 2 Types and sites of protein phosphorylation
| 蛋白质磷酸化类型 | 蛋白质磷酸化位点 | 化学键 |
|---|---|---|
| O-磷酸化 | 苏氨酸(Thr) | P-O |
| 丝氨酸(Ser) | ||
| 酪氨酸(Tyr) | ||
| N-磷酸化 | 组氨酸(His) | P-N |
| 精氨酸(Arg) | ||
| 赖氨酸(Lys) | ||
| S-磷酸化 | 谷氨酸(Glu) | P-O |
| 天冬氨酸(Asp) | ||
| 酰基磷酸化 | 半胱氨酸(Cys) | P-S |
| 蛋白 | 登记号 | 修饰位点 | 参考文献 |
|---|---|---|---|
| AGPS2 | BGIOSGA027135 | S13 (i&j)、S17 (i)、S22 (i)、S35 (i)和S36 (i) | Qiu et al., |
| AGPL2 | BGIOSGA004052 | S62 (i)、S381 (i)和T68 (i&j) | Akihiro et al., |
| GBSSI | BGIOSGA022241 | T57 (j)、Y183 (j)、S283 (j)、T298 (j)、T349 (j)、 S358 (j)、P/S415 (j)、S526 (j)和S569 (j) | Zhang et al., |
| SSIIa | BGIOSGA022586 | S126 (i) | Pang et al., |
| SSIIIa | BGIOSGA028122 | S96 (i&j) | Akihiro et al., |
| BEI | BGIOSGA020506 | S562 (i)、S620 (i)、S814 (i)和S815 (i) | Pang et al., |
| BEIIb | BGIOSGA006344 | S685 (i)和S715 (i) | Pang et al., |
| PUL | BGIOSGA015875 | S154 (i)、S155 (i)和S869 (i) | Pang et al., |
| Pho1 | BGIOSGA009780 | S494 (i)、S645 (i)和S124 (j) | Qiu et al., |
表3 基于磷酸化蛋白质组学鉴定到的水稻胚乳淀粉合成相关蛋白磷酸化位点
Table 3 Identified phosphoproteins involving in starch biosynthesis in rice endosperm based on phosphoproteomics
| 蛋白 | 登记号 | 修饰位点 | 参考文献 |
|---|---|---|---|
| AGPS2 | BGIOSGA027135 | S13 (i&j)、S17 (i)、S22 (i)、S35 (i)和S36 (i) | Qiu et al., |
| AGPL2 | BGIOSGA004052 | S62 (i)、S381 (i)和T68 (i&j) | Akihiro et al., |
| GBSSI | BGIOSGA022241 | T57 (j)、Y183 (j)、S283 (j)、T298 (j)、T349 (j)、 S358 (j)、P/S415 (j)、S526 (j)和S569 (j) | Zhang et al., |
| SSIIa | BGIOSGA022586 | S126 (i) | Pang et al., |
| SSIIIa | BGIOSGA028122 | S96 (i&j) | Akihiro et al., |
| BEI | BGIOSGA020506 | S562 (i)、S620 (i)、S814 (i)和S815 (i) | Pang et al., |
| BEIIb | BGIOSGA006344 | S685 (i)和S715 (i) | Pang et al., |
| PUL | BGIOSGA015875 | S154 (i)、S155 (i)和S869 (i) | Pang et al., |
| Pho1 | BGIOSGA009780 | S494 (i)、S645 (i)和S124 (j) | Qiu et al., |
| 蛋白 | Uniprot 数据库登录号 | 酶编号 | 修饰 位点 | 修饰肽 | 参考文献 |
|---|---|---|---|---|---|
| AGPase S1 | Q69T99 | [EC:2.7.7.27] | 203 | MDYQK(ac)FIQAHR | Wang et al., |
| AGPase S2 | P15280 | [EC:2.7.7.27] | 261 | IVEFAEK(ac)PK | Wang et al., |
| 217 | MDYEK(ac)FIQAHR | Wang et al., | |||
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 250 | ASDYGLVK(ac)FDDSGR | Wang et al., |
| 260 | VIAFSEK(ac)PK | Xing et al., | |||
| 310 | DVLLDILK(ac)SK | Wang et al., | |||
| 312 | SK(ac)YAHLQDFGSEILPR | Wang et al., | |||
| GBSSI | Q0DEV5 | [EC:2.4.1.242] | 444 | KFEK(ac)LLK | Meng et al., |
| 452 | SMEEK(ac)YPGK | Meng et al., | |||
| SSI | Q0DEC8 | [EC:2.4.1.21] | 193 | NFANAFYTEK(ac)HIK | Wang et al., |
| SSIVa | Q5JMA0 | [EC:2.4.1.-] | 589 | AQYYGEHDDFK(ac)R | Meng et al., |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 108 | CLIEK(ac)HEGGLEEFSK | Wang et al., |
| 89 | LEEFK(ac)DHFNYR | Meng et al., | |||
| 103 | YLDQK(ac)CLIEK | Meng et al., | |||
| 118 | HEGGLEEFSK(ac)GYLK | Meng et al., | |||
| 164 | DK(ac)FGIWSIK | Meng et al., | |||
| 236 | YVFK(ac)HPR | Meng et al., | |||
| 372 | GYHK(ac)LWDSR | Meng et al., | |||
| 614 | EGNNWSYDK(ac)CR | Meng et al., | |||
| 662 | QIVSDMNEK(ac)DK | Meng et al., | |||
| 697 | VGCDLPGK(ac)YR | Meng et al., | |||
| 809 | GMK(ac)FVFR | Meng et al., | |||
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134 | VVEELAAEQK(ac)PR | Meng et al., |
| 303 | YIFK(ac)HPQPK | Wang et al., | |||
| 587 | WSEK(ac)CVTYAESHDQALVGDK | Wang et al., | |||
| 688 | FIPGNNNSYDK(ac)CR | Wang et al., | |||
| 738 | KHEEDK(ac)MIIFEK | Meng et al., | |||
| 771 | VGCLKPGK(ac)YK | Meng et al., | |||
| ISA3 | Q6K4A4 | [EC:3.2.1.68] | 130 | K(ac)YFGVAEEK | Meng et al., |
| PUL | Q7X834 | [EC:3.2.1.41] | 805 | NEENWHLIK(ac)PR | Meng et al., |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 747 | FEEAK(ac)QLIR | Meng et al., |
| 169 | YGLFK(ac)QCITK | He et al., | |||
| 409 | HMEIIEEIDK(ac)R | He et al., | |||
| 645 | LVNDVGAVVNNDPDVNK(ac)YLK | He et al., | |||
| 818 | MSILNTAGSGK(ac)FSSDR | He et al., | |||
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 216 | YK(ac)HGLFK | Meng et al., |
| 255 | TDVSYPVK(ac)FYGK | Meng et al., | |||
| 451 | YGTEDTSLLK(ac)K | Meng et al., | |||
| 504 | SLEPSVVVEEK(ac)TVSK | Meng et al., | |||
| 594 | FQNK(ac)TNGVTPR | Meng et al., | |||
| 734 | AFATYVQAK(ac)R | Wang et al., | |||
| 846 | AQGK(ac)FVPDPR | Meng et al., | |||
| 913 | DQK(ac)LWTR | Meng et al., | |||
| 928 | MSILNTASSSK(ac)FNSDR | Meng et al., | |||
| PGM | Q9AUQ4 | [EC:5.4.2.2] | 18 | ATTPFDGQK(ac) PGTSGLR | He et al., |
| 69 | YFSK(ac)DAVQIITK | He et al., | |||
| 206 | LMK(ac)TIFDFESIK | He et al., | |||
| 215 | TIFDFESIK(ac)K | He et al., | |||
| 275 | EDFGGGHPDPNLTYAK(ac)ELVDR | He et al., | |||
| 361 | NLNLK(ac)FFEVPTGWK | He et al., | |||
| 506 | DPVDGSVSK(ac)HQGVR | He et al., | |||
| 543 | VYIEQYEK(ac)DSSK | Meng et al., |
表4 水稻种子淀粉合成蛋白赖氨酸乙酰化修饰
Table 4 Lysine acetylation on starch biosynthesis proteins from rice seeds
| 蛋白 | Uniprot 数据库登录号 | 酶编号 | 修饰 位点 | 修饰肽 | 参考文献 |
|---|---|---|---|---|---|
| AGPase S1 | Q69T99 | [EC:2.7.7.27] | 203 | MDYQK(ac)FIQAHR | Wang et al., |
| AGPase S2 | P15280 | [EC:2.7.7.27] | 261 | IVEFAEK(ac)PK | Wang et al., |
| 217 | MDYEK(ac)FIQAHR | Wang et al., | |||
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 250 | ASDYGLVK(ac)FDDSGR | Wang et al., |
| 260 | VIAFSEK(ac)PK | Xing et al., | |||
| 310 | DVLLDILK(ac)SK | Wang et al., | |||
| 312 | SK(ac)YAHLQDFGSEILPR | Wang et al., | |||
| GBSSI | Q0DEV5 | [EC:2.4.1.242] | 444 | KFEK(ac)LLK | Meng et al., |
| 452 | SMEEK(ac)YPGK | Meng et al., | |||
| SSI | Q0DEC8 | [EC:2.4.1.21] | 193 | NFANAFYTEK(ac)HIK | Wang et al., |
| SSIVa | Q5JMA0 | [EC:2.4.1.-] | 589 | AQYYGEHDDFK(ac)R | Meng et al., |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 108 | CLIEK(ac)HEGGLEEFSK | Wang et al., |
| 89 | LEEFK(ac)DHFNYR | Meng et al., | |||
| 103 | YLDQK(ac)CLIEK | Meng et al., | |||
| 118 | HEGGLEEFSK(ac)GYLK | Meng et al., | |||
| 164 | DK(ac)FGIWSIK | Meng et al., | |||
| 236 | YVFK(ac)HPR | Meng et al., | |||
| 372 | GYHK(ac)LWDSR | Meng et al., | |||
| 614 | EGNNWSYDK(ac)CR | Meng et al., | |||
| 662 | QIVSDMNEK(ac)DK | Meng et al., | |||
| 697 | VGCDLPGK(ac)YR | Meng et al., | |||
| 809 | GMK(ac)FVFR | Meng et al., | |||
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134 | VVEELAAEQK(ac)PR | Meng et al., |
| 303 | YIFK(ac)HPQPK | Wang et al., | |||
| 587 | WSEK(ac)CVTYAESHDQALVGDK | Wang et al., | |||
| 688 | FIPGNNNSYDK(ac)CR | Wang et al., | |||
| 738 | KHEEDK(ac)MIIFEK | Meng et al., | |||
| 771 | VGCLKPGK(ac)YK | Meng et al., | |||
| ISA3 | Q6K4A4 | [EC:3.2.1.68] | 130 | K(ac)YFGVAEEK | Meng et al., |
| PUL | Q7X834 | [EC:3.2.1.41] | 805 | NEENWHLIK(ac)PR | Meng et al., |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 747 | FEEAK(ac)QLIR | Meng et al., |
| 169 | YGLFK(ac)QCITK | He et al., | |||
| 409 | HMEIIEEIDK(ac)R | He et al., | |||
| 645 | LVNDVGAVVNNDPDVNK(ac)YLK | He et al., | |||
| 818 | MSILNTAGSGK(ac)FSSDR | He et al., | |||
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 216 | YK(ac)HGLFK | Meng et al., |
| 255 | TDVSYPVK(ac)FYGK | Meng et al., | |||
| 451 | YGTEDTSLLK(ac)K | Meng et al., | |||
| 504 | SLEPSVVVEEK(ac)TVSK | Meng et al., | |||
| 594 | FQNK(ac)TNGVTPR | Meng et al., | |||
| 734 | AFATYVQAK(ac)R | Wang et al., | |||
| 846 | AQGK(ac)FVPDPR | Meng et al., | |||
| 913 | DQK(ac)LWTR | Meng et al., | |||
| 928 | MSILNTASSSK(ac)FNSDR | Meng et al., | |||
| PGM | Q9AUQ4 | [EC:5.4.2.2] | 18 | ATTPFDGQK(ac) PGTSGLR | He et al., |
| 69 | YFSK(ac)DAVQIITK | He et al., | |||
| 206 | LMK(ac)TIFDFESIK | He et al., | |||
| 215 | TIFDFESIK(ac)K | He et al., | |||
| 275 | EDFGGGHPDPNLTYAK(ac)ELVDR | He et al., | |||
| 361 | NLNLK(ac)FFEVPTGWK | He et al., | |||
| 506 | DPVDGSVSK(ac)HQGVR | He et al., | |||
| 543 | VYIEQYEK(ac)DSSK | Meng et al., |
| 蛋白 | Uniprot数据库登录号 | 酶编号 | 修饰位点 | 参考文献 |
|---|---|---|---|---|
| AGPase S2 | P15280 | [EC:2.7.7.27] | 217、261、263、403、447和476 | Meng et al., |
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 37、250、312、449、459和504 | Meng et al., |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 89、103、164、324、372、500、524和697 | Meng et al., |
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134、191和587 | Meng et al., |
| PUL | Q7X834 | [EC:3.2.1.41] | 732 | Meng et al., |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 412和439 | He et al., |
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 259、255、657和734 | Meng et al., |
| PGM | Q9AUQ4 | [EC:5.4.2.2] | 8和568 | He et al., |
表5 水稻种子淀粉合成蛋白赖氨酸琥珀酰化修饰
Table 5 Lysine succinylation on starch biosynthesis proteins from rice seeds
| 蛋白 | Uniprot数据库登录号 | 酶编号 | 修饰位点 | 参考文献 |
|---|---|---|---|---|
| AGPase S2 | P15280 | [EC:2.7.7.27] | 217、261、263、403、447和476 | Meng et al., |
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 37、250、312、449、459和504 | Meng et al., |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 89、103、164、324、372、500、524和697 | Meng et al., |
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134、191和587 | Meng et al., |
| PUL | Q7X834 | [EC:3.2.1.41] | 732 | Meng et al., |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 412和439 | He et al., |
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 259、255、657和734 | Meng et al., |
| PGM | Q9AUQ4 | [EC:5.4.2.2] | 8和568 | He et al., |
| 蛋白 | Uniprot数据库 登录号 | 酶编号 | Khib修饰位点 | Kmal修饰位点 |
|---|---|---|---|---|
| AGPase S1 | Q69T99 | [EC:2.7.7.27] | 442、249、462、234和203 | - |
| AGPase S2 | P15280 | [EC:2.7.7.27] | 102、106、132、217、239、248、261、263、268、285、360、385、403、406、441、447、456、467、476和496 | 106、360和403 |
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 37、74、187、223、250、263、273、286、301、302、310、312、334、364、371、392、425、443、449、459和504 | 250、312、371和449 |
| AGPase L3 | Q688T8 | [EC:2.7.7.27] | 100、194、196、247、299、326、331、369、446、456和470 | - |
| GBSSI | Q0DEV5 | [EC:2.4.1.242] | 181、192、309、381、385、530、538和549 | - |
| SSI | Q0DEC8 | [EC:2.4.1.21] | 193、196、349、357、429、461、467和570 | - |
| SSIIa | Q0DDE3 | [EC:2.4.1.21] | 151、244、346、378和532 | - |
| SSIIIa | Q6Z1D6 | [EC:2.4.1.21] | 228、 649、761、794、808、961、1203和1604 | - |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 62、64、84、89、103、108、118、122、157、164、171、186、215、236、319、324、372、423、500、506、524、540、549、614、662、664、683、689、697、744、775、796和809 | 108、118、506、524、689和809 |
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134、146、158、191、231、268、299、328、386、466、558、564、571、587、603、612、636、677、688、719、738和773 | 719 |
| ISA3 | Q6K4A4 | [EC:3.2.1.68] | 266和269 | - |
| PUL | Q7X834 | [EC:3.2.1.41] | 123、140、163、239、263、274、388、392、488、535、549、573、590、669、682、732、777、796、805、817和832 | 274和871 |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 115、409、425、533、542、595、721和818 | - |
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 134、255、259、277、289、356、381、410、418、429、441、451、471、493、504、590、617、630、636、657、665、681、725、734、738、846、893、904、913、928、940和946 | 259、493和657 |
| PGM | Q9AUQ4 | [EC:5.4.2.2] | - | 54、458和568 |
表6 水稻种子淀粉合成蛋白赖氨酸2-羟基异丁酰化(Khib)和丙二酰化(Kmal) (Meng et al., 2017; Mujahid et al., 2018)
Table 6 Lysine 2-hydroxyisobutyrylation (Khib) and malonylation (Kmal) on starch biosynthesis proteins from rice seeds (Meng et al., 2017; Mujahid et al., 2018)
| 蛋白 | Uniprot数据库 登录号 | 酶编号 | Khib修饰位点 | Kmal修饰位点 |
|---|---|---|---|---|
| AGPase S1 | Q69T99 | [EC:2.7.7.27] | 442、249、462、234和203 | - |
| AGPase S2 | P15280 | [EC:2.7.7.27] | 102、106、132、217、239、248、261、263、268、285、360、385、403、406、441、447、456、467、476和496 | 106、360和403 |
| AGPase L2 | Q7G065 | [EC:2.7.7.27] | 37、74、187、223、250、263、273、286、301、302、310、312、334、364、371、392、425、443、449、459和504 | 250、312、371和449 |
| AGPase L3 | Q688T8 | [EC:2.7.7.27] | 100、194、196、247、299、326、331、369、446、456和470 | - |
| GBSSI | Q0DEV5 | [EC:2.4.1.242] | 181、192、309、381、385、530、538和549 | - |
| SSI | Q0DEC8 | [EC:2.4.1.21] | 193、196、349、357、429、461、467和570 | - |
| SSIIa | Q0DDE3 | [EC:2.4.1.21] | 151、244、346、378和532 | - |
| SSIIIa | Q6Z1D6 | [EC:2.4.1.21] | 228、 649、761、794、808、961、1203和1604 | - |
| BEI | Q0D9D0 | [EC:2.4.1.18] | 62、64、84、89、103、108、118、122、157、164、171、186、215、236、319、324、372、423、500、506、524、540、549、614、662、664、683、689、697、744、775、796和809 | 108、118、506、524、689和809 |
| BEIIb | Q6H6P8 | [EC:2.4.1.18] | 134、146、158、191、231、268、299、328、386、466、558、564、571、587、603、612、636、677、688、719、738和773 | 719 |
| ISA3 | Q6K4A4 | [EC:3.2.1.68] | 266和269 | - |
| PUL | Q7X834 | [EC:3.2.1.41] | 123、140、163、239、263、274、388、392、488、535、549、573、590、669、682、732、777、796、805、817和832 | 274和871 |
| PhoH | Q8LQ33 | [EC:2.4.1.1] | 115、409、425、533、542、595、721和818 | - |
| PhoL | Q9AUV8 | [EC:2.4.1.1] | 134、255、259、277、289、356、381、410、418、429、441、451、471、493、504、590、617、630、636、657、665、681、725、734、738、846、893、904、913、928、940和946 | 259、493和657 |
| PGM | Q9AUQ4 | [EC:5.4.2.2] | - | 54、458和568 |
| 蛋白 | 登记号 | 修饰位点 |
|---|---|---|
| AGPS1 | BGIOSGA030039 | K94、K464和K484 |
| AGPS2 | BGIOSGA027135 | K106、K132、K385、K403、K406、K476和K496 |
| AGPL2 | BGIOSGA004052 | K41、K78、K134、K191、K227、K254、K316、K338、K394、K396、K463、K508和K513 |
| AGPL3 | BGIOSGA017490 | K509 |
| GBSSI | BGIOSGA022241 | K130、K173、K177、K181、K192、K258、K371、K381、K385、K399、K462、K517、K530、K549、K571和K575 |
| BEI | BGIOSGA020506 | K103、K108和K122 |
| BEIIb | BGIOSGA006344 | K134 |
| PUL | BGIOSGA015875 | K230、K330、K431、K736和K884 |
| Pho1 | BGIOSGA009780 | K277、K445和K941 |
表7 水稻种子淀粉合成蛋白赖氨酸泛素化
Table 7 Lysine ubiquitination on starch biosynthesis proteins from rice seeds
| 蛋白 | 登记号 | 修饰位点 |
|---|---|---|
| AGPS1 | BGIOSGA030039 | K94、K464和K484 |
| AGPS2 | BGIOSGA027135 | K106、K132、K385、K403、K406、K476和K496 |
| AGPL2 | BGIOSGA004052 | K41、K78、K134、K191、K227、K254、K316、K338、K394、K396、K463、K508和K513 |
| AGPL3 | BGIOSGA017490 | K509 |
| GBSSI | BGIOSGA022241 | K130、K173、K177、K181、K192、K258、K371、K381、K385、K399、K462、K517、K530、K549、K571和K575 |
| BEI | BGIOSGA020506 | K103、K108和K122 |
| BEIIb | BGIOSGA006344 | K134 |
| PUL | BGIOSGA015875 | K230、K330、K431、K736和K884 |
| Pho1 | BGIOSGA009780 | K277、K445和K941 |
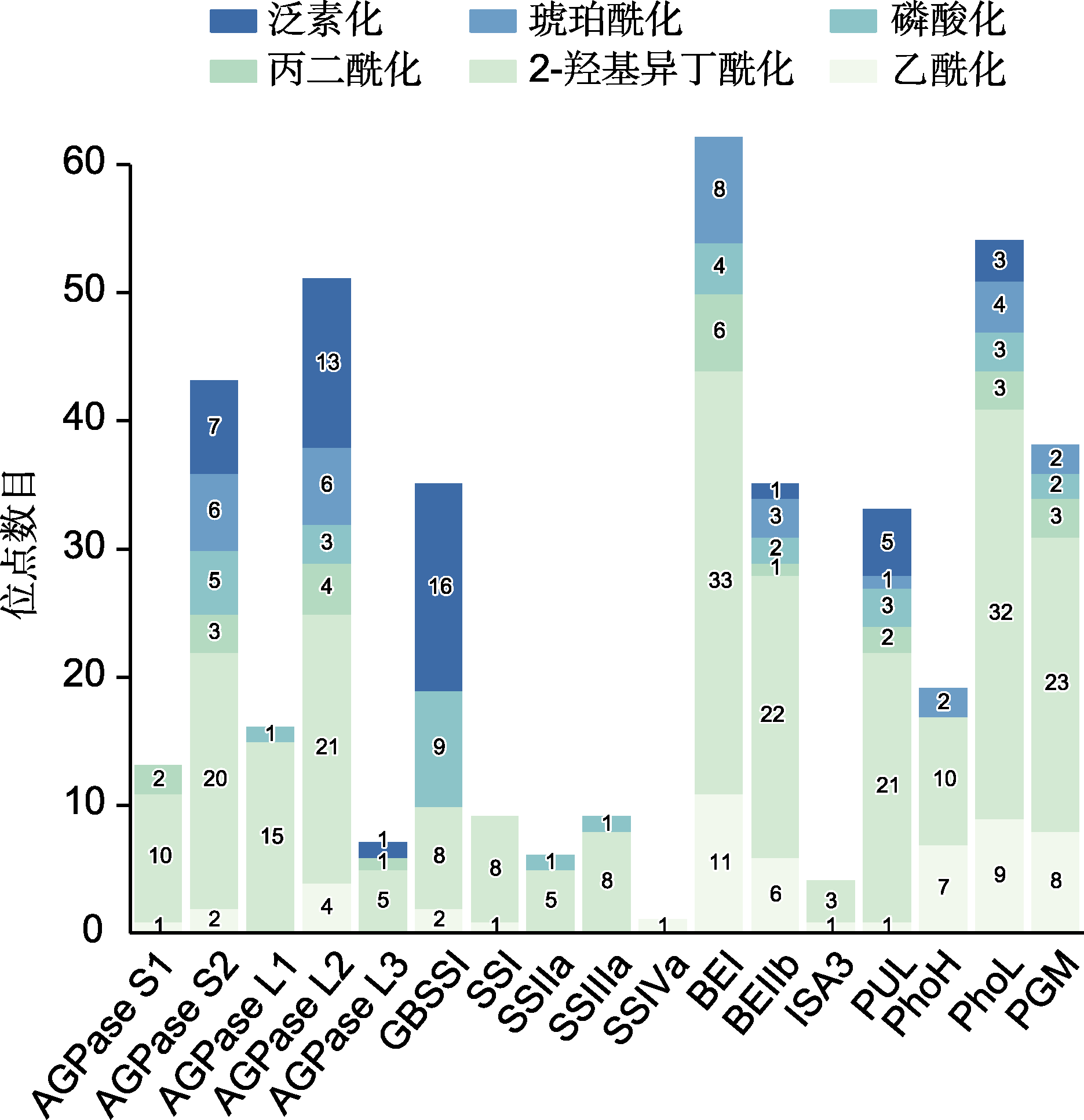
图3 水稻种子中淀粉合成相关酶(SSREs)的蛋白质翻译后修饰(PTMs) 堆积条形图上的数字表示每个蛋白所识别的PTMs修饰位点数量。
Figure 3 Summary of post-translational modifications (PTMs) targeting starch synthesis related enzymes (SSREs) in rice seeds The number of modification site(s) of PTMs identified for each protein are indicated on the stacked bar.
| [1] |
Ahmed Z, Tetlow IJ, Ahmed R, Morell MK, Emes MJ (2015). Protein-protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci 233, 95-106.
DOI PMID |
| [2] |
Akihiro T, Mizuno K, Fujimura T (2005). Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol 46, 937-946.
PMID |
| [3] | Bao JS (2007). Progress in studies on inheritance and improvement of rice starch quality. Mol Plant Breed 5, 1-20. (in Chinese) |
| 包劲松 (2007). 稻米淀粉品质遗传与改良研究进展. 分子植物育种 5, 1-20. | |
| [4] | Bao JS, Deng BW, Zhang L (2023). Molecular and genetic bases of rice cooking and eating quality: an updated review. Cereal Chem 100, 1220-1233. |
| [5] | Bao JS, Kong XL, Xie JK, Xu LJ (2004). Analysis of genotypic and environmental effects on rice starch. 1. Apparent amylose content, pasting viscosity, and gel texture. J Agric Food Chem 52, 6010-6016. |
| [6] | Bao JS, Xia YW (1999). Recent advances on molecular biology of starch biosynthesis in rice (Oryza sativa L.). Chin Bull Bot 16, 352-358. (in Chinese) |
| 包劲松, 夏英武 (1999). 水稻淀粉合成的分子生物学研究进展. 植物学通报 16, 352-358. | |
| [7] | Bao JS, Xu JH (2023). Molecular research for cereal grain quality. Int J Mol Sci 24, 13687. |
| [8] | Bi YD, Wang HX, Lu TC, Li XH, Shen Z, Chen YB, Wang BC (2011). Large-scale analysis of phosphorylated proteins in maize leaf. Planta 233, 383-392. |
| [9] |
Bowman CE, Rodriguez S, Alpergin ESS, Acoba MG, Zhao L, Hartung T, Claypool SM, Watkins PA, Wolfgang MJ (2017). The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem Biol 24, 673-684.
DOI PMID |
| [10] | Cabrera E, Mathews P, Mezhericher E, Beach TG, Deng JJ, Neubert TA, Rostagno A, Ghiso J (2018). Aβ truncated species: implications for brain clearance mechanisms and amyloid plaque deposition. Biochim Biophys Acta Mol Basis Dis 1864, 208-225. |
| [11] |
Chen C, He BS, Liu XX, Ma XD, Liu YJ, Yao HY, Zhang P, Yin JL, Wei X, Koh HJ, Yang C, Xue HW, Fang ZW, Qiao YL (2020). Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa L.). Plant Biotechnol J 18, 83-95.
DOI PMID |
| [12] | Chen GX, Zhou JW, Liu YL, Lu XB, Han CX, Zhang WY, Xu YH, Yan YM (2016). Biosynthesis and regulation of wheat amylose and amylopectin from proteomic and phosphoproteomic characterization of granule-binding proteins. Sci Rep 6, 33111. |
| [13] | Ding JJ, Karim H, Li YL, Harwood W, Guzman C, Lin N, Xu Q, Zhang YZ, Tang HP, Jiang YF, Qi PF, Deng M, Ma J, Wang JR, Chen GY, Lan XJ, Wei YM, Zheng YL, Jiang QT (2021). Re-examination of the APETALA2/Ethy- lene-responsive factor gene family in barley (Hordeum vulgare L.) indicates a role in the regulation of starch synthesis. Front Plant Sci 12, 791584. |
| [14] |
Finkemeier I, Laxa M, Miguet L, Howden AJM, Sweetlove LJ (2011). Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol 155, 1779-1790.
DOI PMID |
| [15] |
Galván-Peña S, Carroll RG, Newman C, Hinchy EC, Palsson-Mcdermott E, Robinson EK, Covarrubias S, Nadin A, James AM, Haneklaus M, Carpenter S, Kelly VP, Murphy MP, Modis LK, O'Neill LA (2019). Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun 10, 338.
DOI PMID |
| [16] | Gao Q, Yan T, Zhang ZJ, Liu SY, Fang XD, Gao DM, Yang YZ, Xu WY, Qiao JH, Cao Q, Ding ZH, Wang Y, Yu JL, Wang XB (2020). Casein kinase 1 regulates cytorhabdovirus replication and transcription by phosphorylating a phosphoprotein serine-rich motif. Plant Cell 32, 2878-2897. |
| [17] | Guo HY, Dong X, An MN, Xia ZH, Wu YH (2024). Research progress in the functions of key enzymes of ubiquitination modification in plant stress responses. Biotechnol Bull 40(4), 1-11. (in Chinese) |
|
郭慧妍, 董雪, 安梦楠, 夏子豪, 吴元华 (2024). 泛素化修饰关键酶在植物抗逆反应中的功能研究进展. 生物技术通报 40(4), 1-11.
DOI |
|
| [18] |
Han C, Yang PF, Sakata K, Komatsu S (2014). Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination. J Proteome Res 13, 1766-1782.
DOI PMID |
| [19] |
He DL, Wang Q, Li M, Damaris RN, Yi XL, Cheng ZY, Yang PF (2016). Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J Proteome Res 15, 879-890.
DOI PMID |
| [20] |
He ZQ, Huang TT, Ao K, Yan XF, Huang Y (2017). Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by ubiquitination. Front Plant Sci 8, 1682.
DOI PMID |
| [21] |
Hennen-Bierwagen TA, Lin QH, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM (2009). Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149, 1541-1559.
DOI PMID |
| [22] |
Hirschey MD, Zhao YM (2015). Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol Cell Proteomics 14, 2308-2315.
DOI PMID |
| [23] | Hu YQ, Zhang YN, Yu SW, Deng GF, Dai GX, Bao JS (2023). Combined effects of BEIIb and SSIIa alleles on amylose contents, starch fine structures and physicochemical properties of indica rice. Foods 12, 119. |
| [24] |
Kosova G, Scott NM, Niederberger C, Prins GS, Ober C (2012). Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet 90, 950-961.
DOI PMID |
| [25] |
Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, Tsai YC, Das S, Chan KC, Zhou M, Andresson T, Weissman AM, Linehan WM, Blair IA, Snyder NW, Meier JL (2017). Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem Biol 24, 231-242.
DOI PMID |
| [26] | Liu DR, Huang WX, Cai XL (2013). Oligomerization of rice granule-bound starch synthase 1 modulates its activity regulation. Plant Sci 210, 141-150. |
| [27] | Liu FS, Ahmed Z, Lee EA, Donner E, Liu Q, Ahmed R, Morell MK, Emes MJ, Tetlow IJ (2012). Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J Exp Bot 63, 1167-1183. |
| [28] | Liu FS, Makhmoudova A, Lee EA, Wait R, Emes MJ, Tetlow IJ (2009). The amylose extender mutant of maize conditions novel protein-protein interactions between star- ch biosynthetic enzymes in amyloplasts. J Exp Bot 60, 4423-4440. |
| [29] |
Luo M, Cheng K, Xu YC, Yang SG, Wu KQ (2017). Plant responses to abiotic stress regulated by histone deacetylases. Front Plant Sci 8, 2147.
DOI PMID |
| [30] | Lv DW, Li X, Zhang M, Gu AQ, Zhen SM, Wang C, Li XH, Yan YM (2014). Large-scale phosphoproteome analysis in seedling leaves of Brachypodium distachyon L. BMC Genomics 15, 375. |
| [31] | Mehrpouyan S, Menon U, Tetlow IJ, Emes MJ (2021). Protein phosphorylation regulates maize endosperm starch synthase IIa activity and protein-protein interactions. Plant J 105, 1098-1112. |
| [32] | Meng XX, Lv YD, Mujahid H, Edelmann MJ, Zhao H, Peng XJ, Peng ZH (2018). Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim Biophys Acta Proteins Proteom 1866, 451-463. |
| [33] |
Meng XX, Mujahid H, Zhang YD, Peng XJ, Redoña ED, Wang CL, Peng ZH (2019). Comprehensive analysis of the lysine succinylome and protein co-modifications in developing rice seeds. Mol Cell Proteomics 18, 2359-2372.
DOI PMID |
| [34] |
Meng XX, Xing SH, Perez LM, Peng XJ, Zhao QY, Redoña ED, Wang CL, Peng ZH (2017). Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in developing rice (Oryza sativa) seeds. Sci Rep 7, 17486.
DOI PMID |
| [35] |
Millar AH, Heazlewood JL, Giglione C, Holdsworth MJ, Bachmair A, Schulze WX (2019). The scope, functions, and dynamics of posttranslational protein modifications. Annu Rev Plant Biol 70, 119-151.
DOI PMID |
| [36] |
Mujahid H, Meng XX, Xing SH, Peng XJ, Wang CL, Peng ZH (2018). Malonylome analysis in developing rice (Oryza sativa) seeds suggesting that protein lysine malonylation is well-conserved and overlaps with acetylation and succinylation substantially. J Proteomics 170, 88-98.
DOI PMID |
| [37] |
Nie LT, Shuai L, Zhu MR, Liu P, Xie ZF, Jiang SW, Jiang HW, Li J, Zhao YM, Li JY, Tan MJ (2017). The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Mol Cell Proteomics 16, 1324-1334.
DOI PMID |
| [38] |
Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56, 3229-3244.
DOI PMID |
| [39] | Pan YH, Chen L, Zhu XY, Li JC, Rashid MAR, Chen C, Qing DJ, Zhou WY, Yang XH, Gao LJ, Zhao Y, Deng GF (2023). Utilization of natural alleles for heat adaptability QTLs at the flowering stage in rice. BMC Plant Biol 23, 256. |
| [40] | Pang YH, Hu YQ, Bao JS (2021). Comparative phosphoproteomic analysis reveals the response of starch metabolism to high-temperature stress in rice endosperm. Int J Mol Sci 22, 10546. |
| [41] | Pang YH, Ying YN, Xu FF, Bao JS (2024). Integrated analysis of phosphoproteome and ubiquitinated proteome in rice endosperm under high temperature stress. J Zhejiang Univ Agric Life Sci 50, 382-392. (in Chinese) |
| 庞悦涵, 应逸宁, 徐非非, 包劲松 (2024). 高温胁迫下水稻胚乳磷酸化与泛素化蛋白质组学联合分析. 浙江大学学报(农业与生命科学版) 50, 382-392. | |
| [42] | Pang YH, Zhou X, Chen YL, Bao JS (2018). Comparative phosphoproteomic analysis of the developing seeds in two indica rice (Oryza sativa L.) cultivars with different starch quality. J Agric Food Chem 66, 3030-3037. |
| [43] | Qiu JH, Hou YX, Tong XH, Wang YF, Lin HY, Liu Q, Zhang W, Li ZY, Nallamilli BR, Zhang J (2016). Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol Biol 90, 249-265. |
| [44] |
Smith-Hammond CL, Swatek KN, Johnston ML, Thelen JJ, Miernyk JA (2014). Initial description of the developing soybean seed protein Lys-Nε-acetylome. J Proteomics 96, 56-66.
DOI PMID |
| [45] |
Svensson K, Labarge SA, Sathe A, Martins VF, Tahvilian S, Cunliffe JM, Sasik R, Mahata SK, Meyer GA, Philp A, David LL, Ward SR, Mccurdy CE, Aslan JE, Schenk S (2020). P300 and cAMP response element-binding protein-binding protein in skeletal muscle homeostasis, contractile function, and survival. J Cachexia Sarcopenia Muscle 11, 464-477.
DOI PMID |
| [46] | Tan MJ, Peng C, Anderson KA, Chhoy P, Xie ZY, Dai LZ, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu GF, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, Mcguire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao YM (2014). Lysine glutarylation is a protein post-translational modification regulated by SIRT5. Cell Metab 19, 605-617. |
| [47] | Tang ZW, Zhang DP (2023). Research progress on the molecular mechanism of starch accumulation in rice endosperm. Chin Bull Bot 58, 612-621. (in Chinese) |
|
唐子雯, 张冬平 (2023). 水稻胚乳淀粉积累过程的分子机理研究进展. 植物学报 58, 612-621.
DOI |
|
| [48] | Teng B, Zeng RZ, Wang YC, Liu ZQ, Zhang ZM, Zhu HT, Ding XH, Li WT, Zhang GQ (2012). Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol Breed 30, 583-595. |
| [49] |
Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu FS, Bresolin NS, Wait R, Morell MK, Emes MJ (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146, 1878-1891.
DOI PMID |
| [50] |
Tetlow IJ, Wait R, Lu ZX, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ (2004). Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16, 694-708.
DOI PMID |
| [51] | Wang YF, Hou YX, Qiu JH, Li ZY, Zhao J, Tong XH, Zhang J (2017). A quantitative acetylomic analysis of early seed development in rice (Oryza sativa L.). Int J Mol Sci 18, 1376. |
| [52] |
Wang ZA, Cole PA (2020). The chemical biology of reversible lysine post-translational modifications. Cell Chem Biol 27, 953-969.
DOI PMID |
| [53] | Wu LP, Meng XX, Huang HZ, Liu YY, Jiang WM, Su XL, Wang ZJ, Meng F, Wang LH, Peng DY, Xing SH (2022). Comparative proteome and phosphoproteome analyses reveal different molecular mechanism between stone plan- ting under the forest and greenhouse planting of Dendrobium huoshanense. Front Plant Sci 13, 937392. |
| [54] | Wu X, Oh MH, Schwarz EM, Larue CT, Sivaguru M, Imai BS, Yau PM, Ort DR, Huber SC (2011). Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol 155, 1769-1778. |
| [55] |
Xie X, Kang HX, Liu WD, Wang GL (2015). Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J Proteome Res 14, 2017-2025.
DOI PMID |
| [56] | Xing SH, Meng XX, Zhou LH, Mujahid H, Zhao CF, Zhang YD, Wang CL, Peng ZH (2016). Proteome profile of starch granules purified from rice (Oryza sativa) endosperm. PLoS One 11, e0168467. |
| [57] | Xiong YH, Peng XJ, Cheng ZY, Liu WD, Wang GL (2016). A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J Pro- teomics 138, 20-29. |
| [58] | Xu M, Tian XM, Ku TT, Wang GY, Zhang EY (2021). Global identification and systematic analysis of lysine malonylation in maize (Zea mays L.). Front Plant Sci 12, 728338. |
| [59] | Xu Y, Li XX, Liang WX, Liu MJ (2020). Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in the phytopathogenic fungus Botrytis cinerea. Front Microbiol 11, 585614. |
| [60] | Xue C, Liu S, Chen C, Zhu J, Yang XB, Zhou Y, Guo R, Liu XY, Gong ZY (2018). Global proteome analysis links lysine acetylation to diverse functions in Oryza sativa. Pro- teomics 18, 1700036. |
| [61] | Xue C, Qiao ZY, Chen X, Cao PH, Liu K, Liu S, Ye L, Gong ZY (2020). Proteome-wide analyses reveal the diverse functions of lysine 2-hydroxyisobutyrylation in Oryza sativa. Rice 13, 34. |
| [62] | Xue H, Zhang QF, Wang PQ, Cao BJ, Jia CC, Cheng B, Shi YH, Guo WF, Wang ZL, Liu ZX, Cheng H (2022). QPTMplants: an integrative database of quantitative post- translational modifications in plants. Nucleic Acids Res 50, D1491-D1499. |
| [63] | Yang WQ, Zhang W, Wang XX (2017). Post-translational control of ABA signaling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J 15, 4-14. |
| [64] | Yin XJ, Wang X, Komatsu S (2018). Phosphoproteomics: protein phosphorylation in regulation of seed germination and plant growth. Curr Protein Pept Sci 19, 401-412. |
| [65] | Ying YN, Hu YQ, Zhang YN, Tappiban P, Zhang ZW, Dai GX, Deng GF, Bao JS, Xu FF (2023a). Identification of a new allele of soluble starch synthase IIIa involved in the elongation of amylopectin long chains in a chalky rice mu- tant. Plant Sci 328, 111567. |
| [66] | Ying YN, Pang YH, Bao JS (2023b). Comparative ubiquitome analysis reveals diverse functions of ubiquitination in rice seed development under high-temperature stress. Seed Biol 2, 23. |
| [67] | Ying YN, Xu FF, Zhang ZW, Tappiban P, Bao JS (2022). Dynamic change in starch biosynthetic enzymes complexes during grain-filling stages in BEIIb active and deficient rice. Int J Mol Sci 23, 10714. |
| [68] | Yu GW, Gaoyang YZ, Liu L, Shoaib N, Deng YW, Zhang N, Li YP, Huang YB (2022). The structure, function, and regulation of starch synthesis enzymes SSIII with emphasis on maize. Agronomy 12, 1359. |
| [69] | Yu GW, Lv YA, Shen LY, Wang YB, Qing Y, Wu N, Li YP, Huang HH, Zhang N, Liu YH, Hu YF, Liu HM, Zhang JJ, Huang YB (2019). The proteomic analysis of maize endosperm protein enriched by phos-tagtm reveals the phosphorylation of Brittle-2 subunit of ADP-Glc pyrophosphorylase in starch biosynthesis process. Int J Mol Sci 20, 986. |
| [70] |
Yu HT, Wang T (2016). Proteomic dissection of endosperm starch granule associated proteins reveals a network coordinating starch biosynthesis and amino acid metabolism and glycolysis in rice endosperms. Front Plant Sci 7, 707.
DOI PMID |
| [71] | Zhang CQ, Zhu JH, Chen SJ, Fan XL, Li QF, Lu Y, Wang M, Yu HX, Yi CD, Tang SZ, Gu MH, Liu QQ (2019a). Wxlv, the ancestral allele of rice Waxy gene. Mol Plant 12, 1157-1166. |
| [72] | Zhang K, Xiong YH, Sun WX, Wang GL, Liu WD (2019b). Global proteomic analysis reveals widespread lysine succinylation in rice seedlings. Int J Mol Sci 20, 5911. |
| [73] | Zhang M, Lv DW, Ge P, Bian YW, Chen GX, Zhu GR, Li XH, Yan YM (2014a). Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J Proteomics 109, 290-308. |
| [74] | Zhang M, Ma CY, Lv DW, Zhen SM, Li XH, Yan YM (2014b). Comparative phosphoproteome analysis of the developing grains in bread wheat (Triticum aestivum L.) under well-watered and water-deficit conditions. J Proteome Res 13, 4281-4297. |
| [75] |
Zhang YM, Song LM, Liang WX, Mu P, Wang S, Lin Q (2016). Comprehensive profiling of lysine acetylproteome analysis reveals diverse functions of lysine acetylation in common wheat. Sci Rep 6, 21069.
DOI PMID |
| [76] |
Zhang ZH, Tan MJ, Xie ZY, Dai LZ, Chen Y, Zhao YM (2011). Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol 7, 58-63.
DOI PMID |
| [77] | Zhang ZW, Ying YN, Zhang L, Dai GX, Deng GF, Bao JS, Xu FF (2023). Starch structural reasons for the effects of SSIIIa deficiency on the textural and digestive properties of cooked rice. J Cereal Sci 111, 103671. |
| [78] | Zhang ZX, Zhao H, Tang J, Li Z, Li Z, Chen DM, Lin WX (2014c). A proteomic study on molecular mechanism of poor grain-filling of rice (Oryza sativa L.) inferior spikelets. PLoS One 9, e89140. |
| [79] | Zhou X (2020). Genetic and Molecular Structural Basis for Resistant Starch and Other Functional Properties in Rice (Oryza sativa L.). PhD dissertation. Hangzhou: Zhejiang University. pp. 1-134. (in Chinese) |
| 周鑫 (2020). 稻米淀粉消化特性等功能性状的遗传与结构基础研究. 博士论文. 杭州: 浙江大学. pp. 1-134. | |
| [80] |
Zhu LY, Cheng H, Peng GQ, Wang SS, Zhang ZG, Ni ED, Fu XD, Zhuang CX, Liu ZX, Zhou H (2020). Ubiquitinome profiling reveals the landscape of ubiquitination regulation in rice young panicles. Genom Proteom Bioinf 18, 305-320.
DOI PMID |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 周婧, 高飞. 植物缺铁诱导型香豆素合成及其在铁吸收中的功能研究进展[J]. 植物学报, 2025, 60(3): 460-471. |
| [3] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 徐秀苹, 杨小雨, 冯旻. 成熟禾谷类作物种子胚乳细胞的显微CT扫描方法[J]. 植物学报, 2025, 60(1): 81-89. |
| [6] | 胡海涛, 武越, 杨玲. 植物NAD(P)+的生物合成及其生物学功能研究进展[J]. 植物学报, 2025, 60(1): 114-131. |
| [7] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [8] | 蔡慧颖, 李兰慧, 林阳, 梁亚涛, 杨光, 孙龙. 白桦叶片和细根非结构性碳水化合物对火后时间的响应[J]. 植物生态学报, 2024, 48(6): 780-793. |
| [9] | 范雪兰, 落艳娇, 徐超群, 郭宝林. 淫羊藿类黄酮生物合成相关基因研究进展[J]. 植物学报, 2024, 59(5): 834-846. |
| [10] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [11] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [12] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [13] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [14] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [15] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||