

植物学报 ›› 2016, Vol. 51 ›› Issue (2): 184-193.DOI: 10.11983/CBB15080 cstr: 32102.14.CBB15080
收稿日期:2015-05-18
接受日期:2015-12-13
出版日期:2016-03-01
发布日期:2016-03-31
通讯作者:
E-mail: 基金资助:
Kaijian Lei1,2, Jing Ren1, Yuanyuan Zhu1, Guoyong An1,*( )
)
Received:2015-05-18
Accepted:2015-12-13
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: 摘要: 根际酸化是植物适应低磷胁迫的重要策略, 但植物是如何感知和转导低磷信号, 进而促进根际酸化的分子机制至今还不十分清楚。利用pH指示剂(溴甲酚紫)显色法从拟南芥(Arabidopsis thaliana) T-DNA插入突变体库中分离得到了1株低磷诱导根际酸化缺失突变体spl1。在含溴甲酚紫的低磷培养基上培养8小时, 野生型拟南芥根际培养基的颜色变为黄色, 而突变体spl1根际培养基的颜色没有明显变化, 表明spl1的低磷根际酸化反应能力降低。当低磷胁迫处理延长20天, spl1叶片的花青素积累明显高于野生型。同时也出现, 即使在磷营养正常条件下, spl1突变体也表现出根毛数量与长度增加的特征。进一步的研究表明, 在低磷条件下, spl1突变体根部的磷含量略高于野生型, 与磷转运相关基因的表达量明显高于野生型。分子遗传学分析结果表明, SPL1基因受低磷胁迫诱导, 主要在拟南芥的叶片和花等组织中表达, 其编码的蛋白广泛分布在细胞的各个部位。以上结果表明, SPL1参与介导低磷诱导的拟南芥根际酸化反应, 调节多种低磷胁迫反应及低磷条件下磷饥饿诱导基因的表达。
雷凯健, 任晶, 朱园园, 安国勇. 拟南芥SPL1基因参与调节低磷条件下的根际酸化反应. 植物学报, 2016, 51(2): 184-193.
Kaijian Lei, Jing Ren, Yuanyuan Zhu, Guoyong An. SPL1 is Involved in the Regulation of Rhizosphere Acidification Reaction Under Low Phosphate Condition in Arabidopsis. Chinese Bulletin of Botany, 2016, 51(2): 184-193.
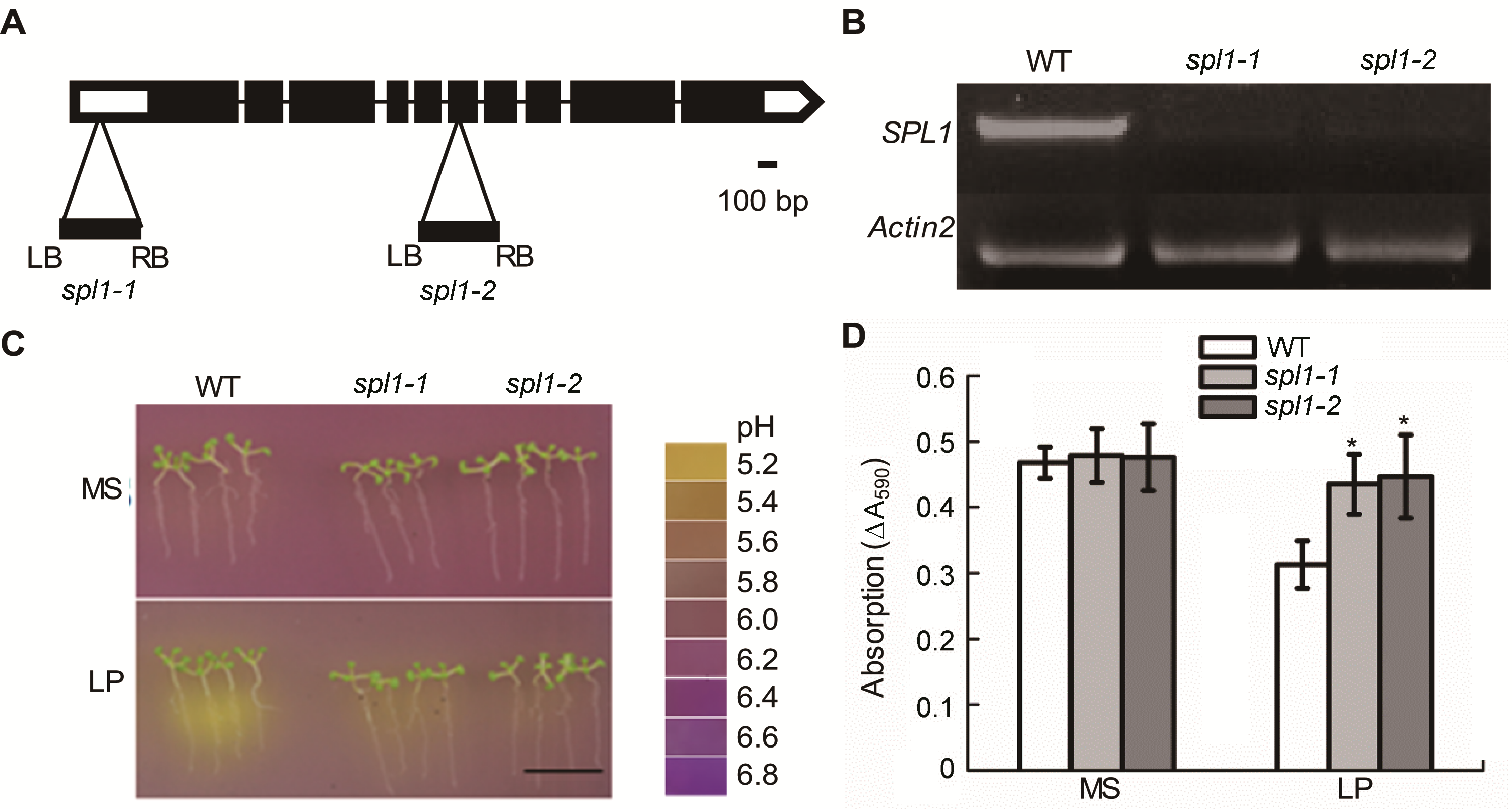
图1 拟南芥spl1突变体的鉴定及根际酸化缺失表型分析 (A) spl1-1和spl1-2的T-DNA插入位点示意图; (B) RT-PCR分析SPL1的表达; (C) 溴甲酚紫显色法分析WT和spl1在MS及低磷(LP)培养基上(12.5 μmol∙L−1 H2PO4−)的根际酸化反应(Bar=1 cm); (D) WT和spl1的酸化定量。*表示与野生型相比差异显著(P<0.05)。
Figure 1 Arabidopsis thaliana spl1 mutants show the phenotype of rhizosphere acidification deficiency and decreased acidi- fication capacity (A) T-DNA insertion sites in spl1-1 and spl1-2; (B) Reverse transcriptase-PCR analysis of SPL1 expression; (C) The rhizosphere acidification reaction of WT and spl1 mutants under MS and low phosphate (LP) conditions (12.5 μmo∙L−1 H2PO4−) (Bar=1 cm); (D) The acidification capacity of WT and spl1 mutants. Asterisk represent statistically differences compared with the wild type (P<0.05).
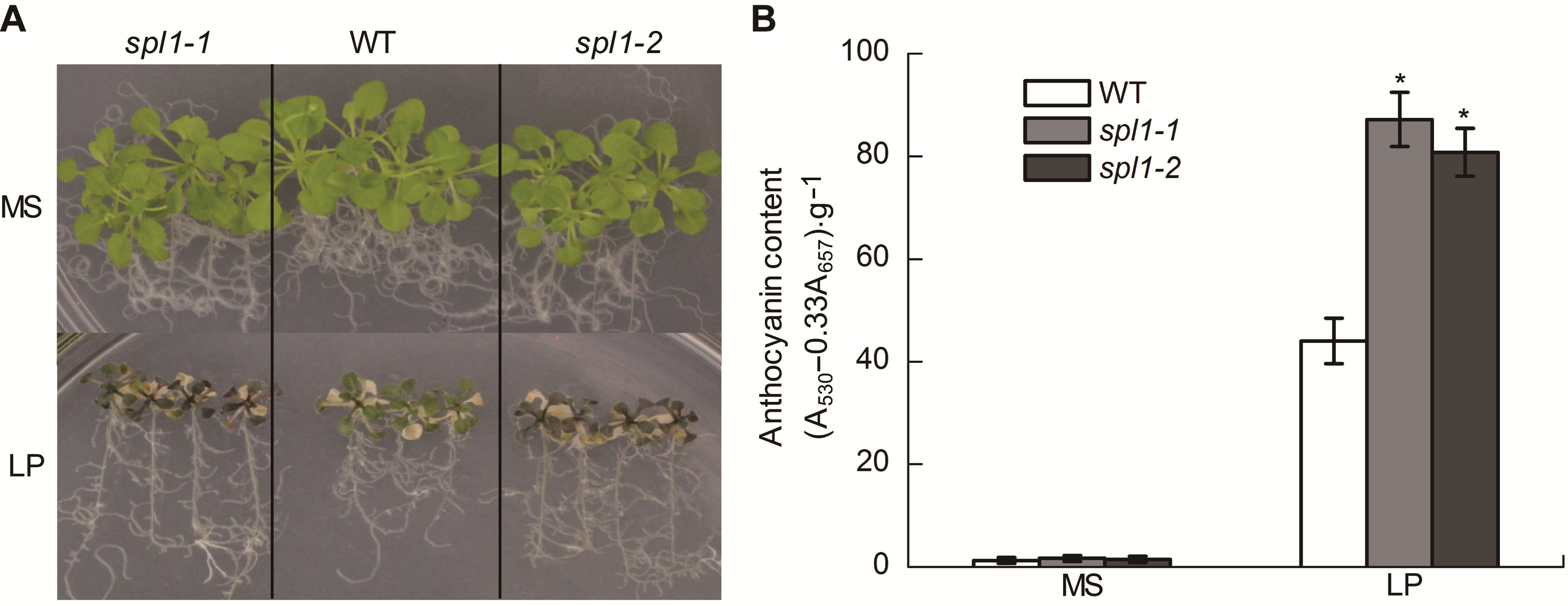
图2 低磷条件下拟南芥突变体spl1-1和spl1-2花青素的积累 (A) 在正常和低磷条件下, WT、spl1-1和spl1-2的叶片表型特征; (B) 低磷处理20天, 野生型(WT)、spl1-1和spl1-2的花青素含量变化。*表示与野生型相比差异显著(P<0.05)。
Figure 2 The anthocyanin accumulation in Arabidopsis thaliana spl1-1 and spl1-2 under low Pi condition (A) The anthocyanin accumulation in wild type (WT), spl1-1, and spl1-2 plants during Pi sufficient and Pi deprivation; (B) Anthocyanin content was determined in WT and spl1 plants grown with normal and low Pi (LP) on the twentieth day of Pi starvation. Asterisk represents statistically differences compared with the wild type (P<0.05).
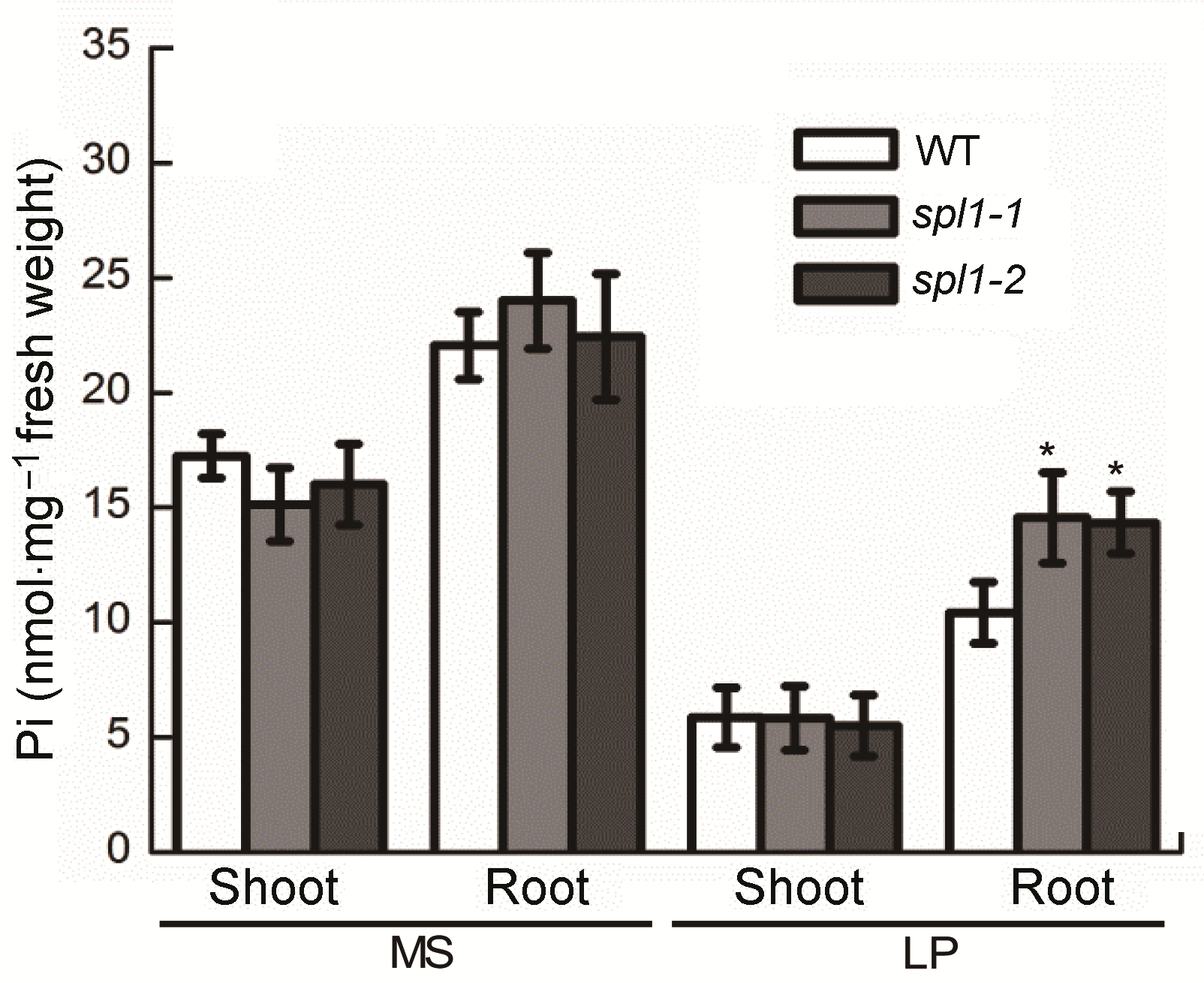
图3 不同条件下拟南芥突变体spl1的无机磷含量变化 生长7天的拟南芥幼苗移植到MS或低磷(LP)培养基上处理14天, 然后测定无机磷含量。*表示与野生型相比差异显著(P<0.05)。
Figure 3 The Pi content analysis of Arabidopsis thaliana spl1 mutants Seven-day-old seedlings were transferred to MS or low Pi (LP) medium for 14 d and then harvested for Pi content analy- sis. Asterisk represents statistically differences compared with the wild type (P<0.05).
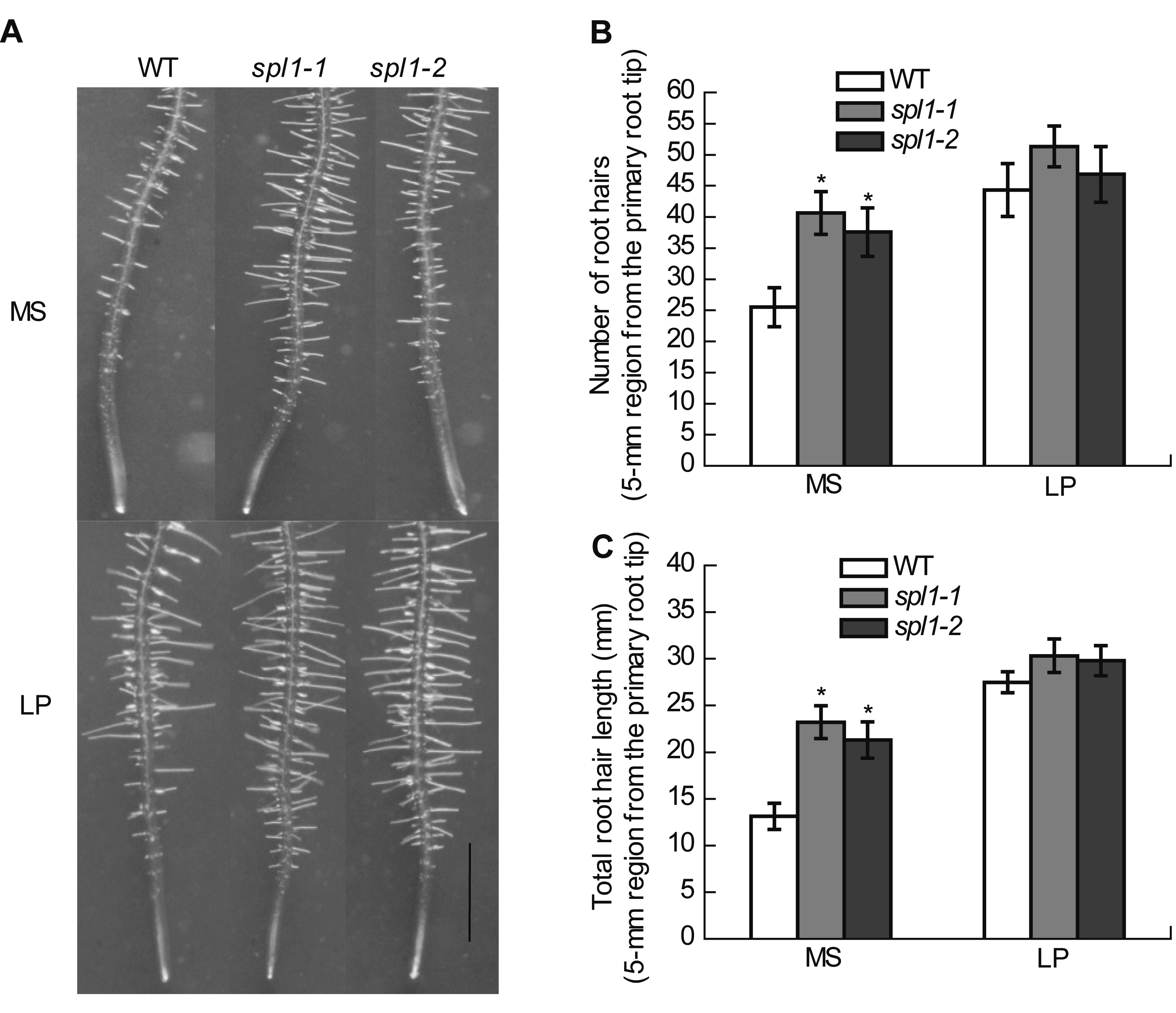
图4 SPL1突变对根毛形态特征的影响 (A) 不同磷营养条件下, WT、spl1-1和spl1-2的根毛表型特征(Bar=1 mm); (B), (C) 在MS及低磷(LP)条件下的根毛数目(B)及根毛长度(C)统计结果(P<0.05)。*表示与对照相比差异显著。
Figure 4 SPL1 alters root hair architecture (A) Images of root tips with intact root hair from WT, spl1-1 and spl1-2 plants (Bar=1 mm); (B), (C) Number of root hair (B) and root hair length (C) under MS and low Pi (LP) condition. *Data significantly different from the corresponding controls are indicated (P<0.05).
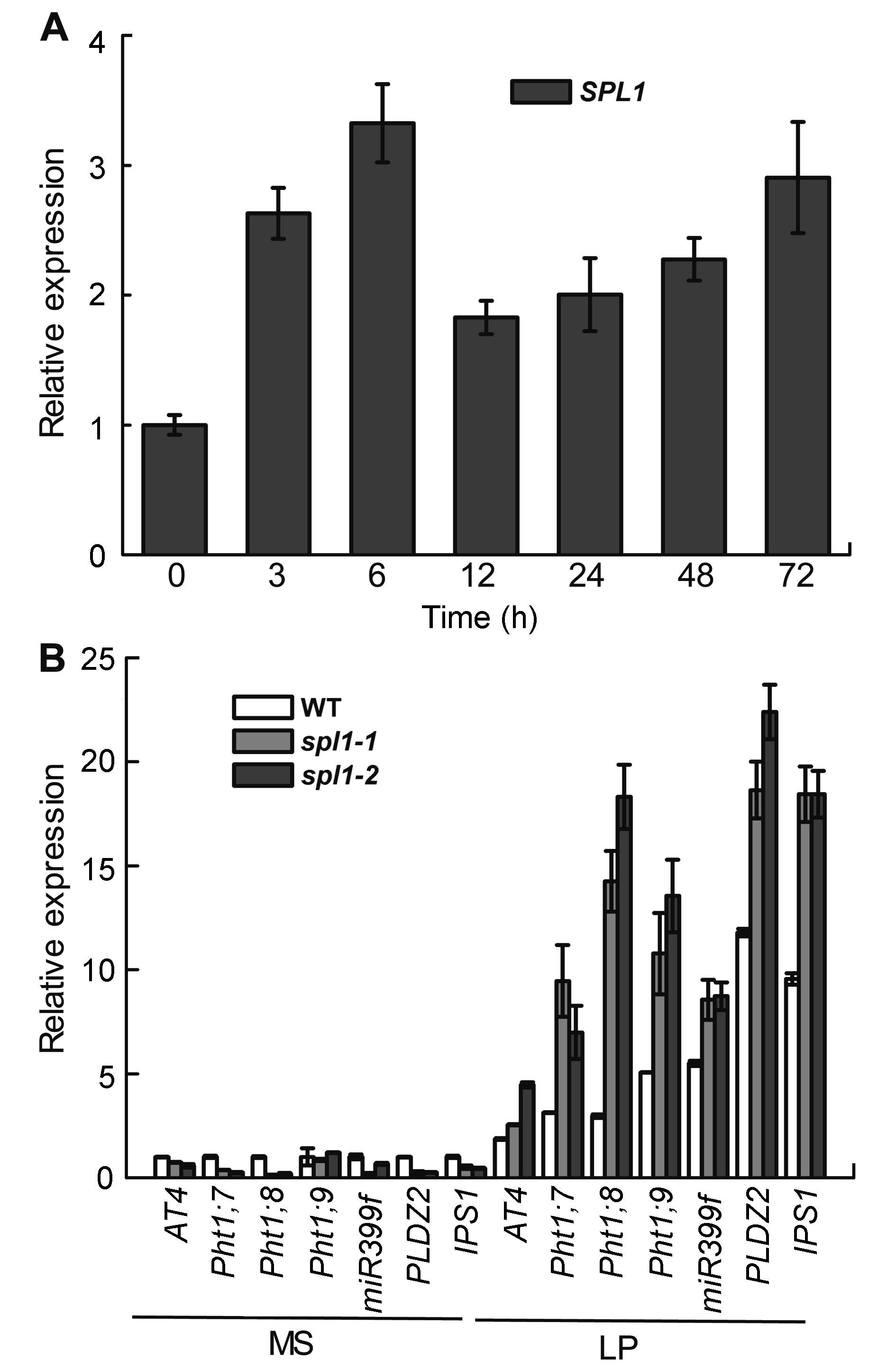
图5 低磷对SPL1基因表达的影响(A)及磷饥饿诱导基因在拟南芥spl1突变体中的表达(B)
Figure 5 Expression analyses of the SPL1 gene (A) in Arabidopsis seedlings in response to low phosphate (LP) stress and expression pattern of Pi starvation induced genes in spl1 mutants (B)
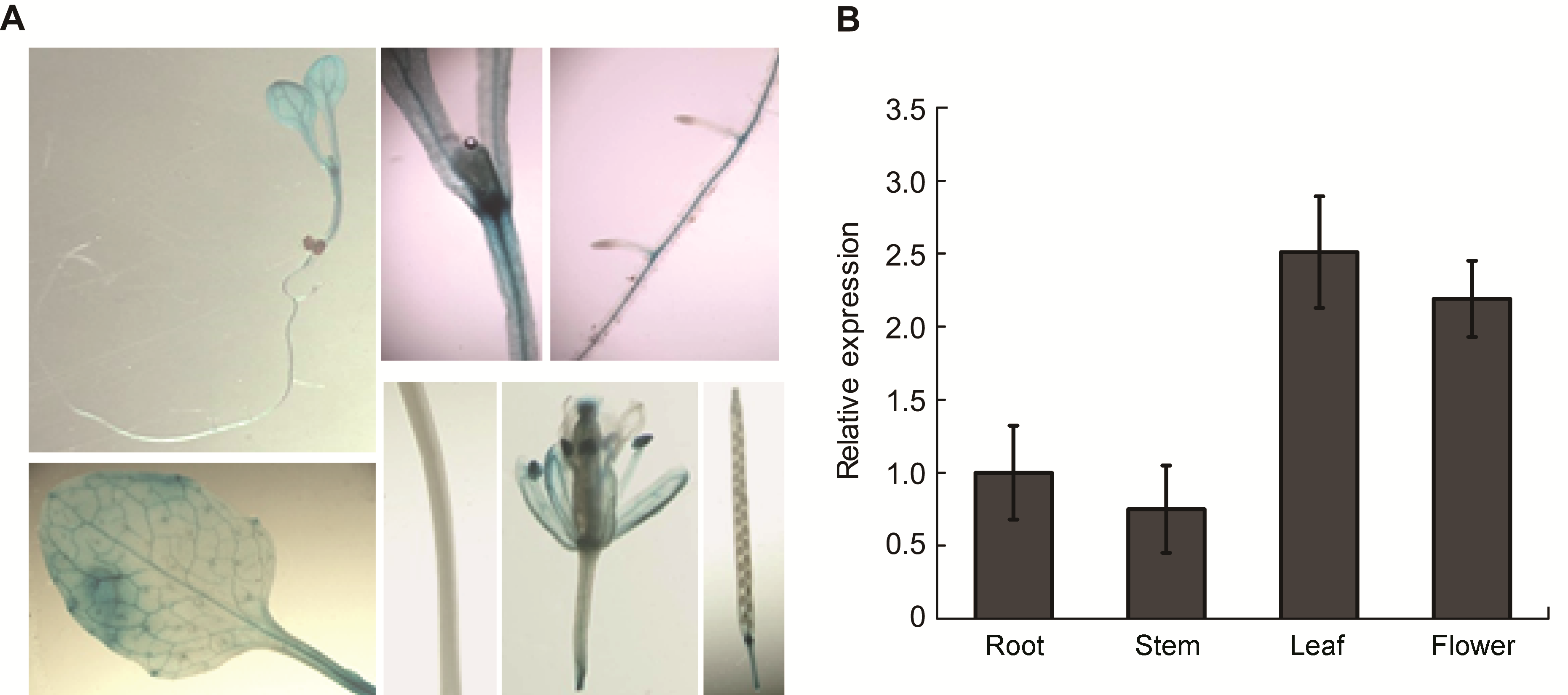
图6 SPL1基因的组织表达特性 (A) SPL1::GUS转基因植株的GUS染色; (B) 用qRT-PCR方法分析SPL1基因在根、茎、叶和花中的组织表达
Figure 6 Tissue-specific expression of SPL1 (A) Histochemical localization of GUS activity directed by SPL1::GUS fusions in transgenic Arabidopsis; (B) qRT-PCR showed the relative abundant of SPL1 gene in different tissues, including root, stem, leaf and flower
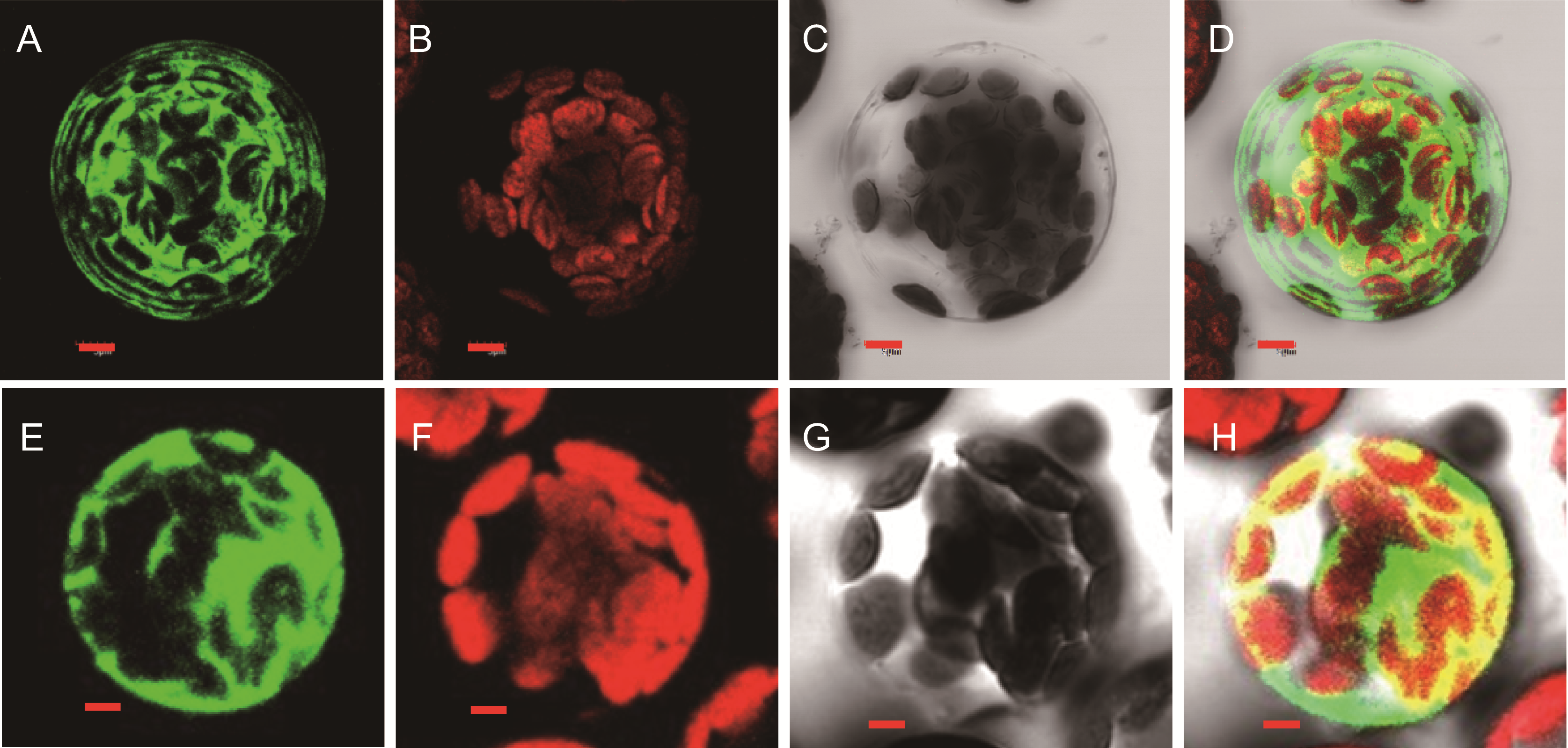
图7 pHBT (A−D)及SPL1 (E−H)蛋白的细胞定位特性 (A), (E) 绿色荧光图; (B), (F) 叶绿体自发荧光图; (C), (G) 透射光图; (D), (H) 叠加图。Bar=5 µm
Figure 7 Localization of GFP signals from pHBT empty vector (A−D) and SPL1 (E−H) fused with GFP (A), (E) Fluorescence images under confocal microscopy; (B), (F) Chloroplast fluorescence; (C), (G) Bright-field images of the cells; (D), (H) Merged fluorescence. Bar=5 µm
| [1] | 雷凯健, 安国勇 (2014). 植物miRNA介导磷信号转导的研究进展. 植物生理学报 50, 1071-1078. |
| [2] | 张健, 徐金相, 孔英珍, 纪振动, 王兴春, 安丰英, 李超, 孙加强, 张素芝, 杨晓辉, 牟金叶, 刘新仿, 李家洋, 薛勇彪, 左建儒 (2005). 化学诱导激活型拟南芥突变体库的构建及分析. 遗传学报 32, 1082-1088. |
| [3] | Ames BN (1966). Assay of inorganic phosphate, total phosphate and phosphatase. Methods Enzymol 8, 115-118. |
| [4] | Cordell D, Dranger JO, White S (2009). The story of phosphorus: global food security and food for thought. Global Environ Change 19, 292-305. |
| [5] |
Cruz-Ramirez A, Oropeza-Aburto A, Razo-Hernandez F, Ramirez-Chavez E, Herrera-Estrella L (2006). Phos- pholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103, 6765-6770.
DOI PMID |
| [6] | Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80, 1108-1117. |
| [7] |
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39, 1033-1037.
DOI PMID |
| [8] |
Gomes-Junior RA, Moldes CA, Delite FS, Pompeu GB, Gratão PL, Mazzafera P, Lea PJ, Azevedo RA (2006). Antioxidant metabolism of coffee cell suspension cultures in response to cadmium. Chemosphere 65, 1330-1337.
DOI PMID |
| [9] | Gou JY, Felippes F, Liu CJ, Weigel D, Wang JW (2011). Negative regulation of anthocyanin biosynthesis in Ara- bidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512-1522. |
| [10] |
Hinsinger P, Gobran GR, Gregory PJ, Wenzel WW (2005). Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytol 168, 293-303.
DOI PMID |
| [11] |
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009). Uncovering small RNA-media- ted responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151, 2120-2132.
DOI PMID |
| [12] |
Jefferson RA, Kavanagh TA, Bevan MW (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6, 3901-3907.
DOI PMID |
| [13] |
Khorassani R, Hettwer U, Ratzinger A, Steingrobe B, Karlovsky P, Claassen N (2011). Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol 11, 121-128.
DOI PMID |
| [14] | Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduc- tion. Plant Cell 15, 2399-2407. |
| [15] | Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient tem- perature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol 159, 461-478. |
| [16] |
Lapis-Gaza HR, Jost R, Finnegan PM (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14, 334.
DOI PMID |
| [17] | Lei KJ, Lin YM, Ren J, Bai L, Miao YC, An GY, Song CP (2016). Modulation of the phosphate-deficient responses by the microRNA156 and its targeted SQUAMOSA PRO- MOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol 57, 192-203. |
| [18] | Lei KJ, Xie JY, Zhu YY, Song CP, An GY (2015). Screen- ing and analysis of rhizosphere acidification deficiency mutants in Arabidopsis thaliana under low phosphorus. Soil Sci Plant Nutr 61, 493-500. |
| [19] | Li M, Shinano T, Tadano T (1997). Distribution of exu- dates of lupin roots in the rhizosphere under phosphor- rus deficient conditions. Soil Sci Plant Nutr 43, 237-245. |
| [20] | Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147, 732-746. |
| [21] |
López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003). The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6, 280-287.
DOI PMID |
| [22] | Neumann G, Römheld V (1999). Root excretion of carbo- xylic acids and protons in phosphorus-deficient plants. Plant Soil 211, 121-130. |
| [23] | Raghothama KG (1999). Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50, 665-693. |
| [24] |
Rausch C, Bucher M (2002). Molecular mechanisms of phosphate transport in plants. Planta 216, 23-37.
DOI PMID |
| [25] |
Rouached H, Arpat AB, Poirier Y (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3, 288-299.
DOI PMID |
| [26] | Santi S, Schmidt W (2009). Dissecting iron deficiency- induced proton extrusion in Arabidopsis roots. New Phytol 183, 1072-1084. |
| [27] | Shin H, Shin HS, Chen R, Harrison MJ (2006). Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45, 712-726. |
| [28] |
Sternberg D, Mandels GR (1979). Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol 139, 761-769.
DOI PMID |
| [29] | Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcript- tion factors. Plant Cell 26, 1792-1807. |
| [30] | Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acqui- sition in Arabidopsis. Plant Physiol 67, 1579-1591. |
| [31] |
Ticconi CA, Abel S (2004). Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9, 548-555.
PMID |
| [32] | Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P (2003). SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15, 1009-1019. |
| [33] |
Vance CP, Uhde-Stone C, Allan DL (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157, 423-447.
DOI PMID |
| [34] | Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008). Dual effects of miR156-targeted SPL genes and CYP- 78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231-1243. |
| [35] |
Watt M, Evans JR (1999). Proteoid roots. Physiology and development. Plant Physiol 121, 317-323.
PMID |
| [36] |
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750-759.
DOI PMID |
| [37] | Xing SP, Salinas M, Höhmann S, Berndtgen R, Huijser P (2010). MiR156-targeted and non targeted SBP-box trans- cription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935-3950. |
| [38] |
Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009). SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 is a central regulator for copper homeo- stasis in Arabidopsis. Plant Cell 21, 347-361.
DOI PMID |
| [39] | Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTERBIN- DING-LIKE transcription factors. Plant Cell 24, 3320-3332. |
| [40] |
Zhang Y, Schwarz S, Saedler H, Huijser P (2007). SPL8, a local regulator in a subset of gibberellins mediated developmental processes in Arabidopsis. Plant Mol Biol 63, 429-439.
DOI PMID |
| [41] |
Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, Li X, Zhang Y (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107, 18220-18225.
DOI PMID |
| [1] | 刘雨函, 曹启江, 张诗晗, 李益慧, 王菁, 谭晓萌, 刘筱儒, 王显玲. 拟南芥AtFTCD-L参与根系响应土壤紧实度的机制研究[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 景艳军, 林荣呈. 蓝光受体CRY2化身“暗黑舞者”[J]. 植物学报, 2024, 59(6): 878-882. |
| [3] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [4] | 陈艳晓, 李亚萍, 周晋军, 解丽霞, 彭永彬, 孙伟, 和亚男, 蒋聪慧, 王增兰, 郑崇珂, 谢先芝. 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024, 59(3): 481-494. |
| [5] | 杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础[J]. 植物学报, 2024, 59(3): 373-382. |
| [6] | 王钢, 王二涛. “卫青不败由天幸”——WeiTsing的广谱抗根肿病机理被揭示[J]. 植物学报, 2023, 58(3): 356-358. |
| [7] | 杨永青, 郭岩. 植物细胞质外体pH感受机制的解析[J]. 植物学报, 2022, 57(4): 409-411. |
| [8] | 支添添, 周舟, 韩成云, 任春梅. PAD4突变加速拟南芥酪氨酸降解缺陷突变体sscd1的程序性细胞死亡[J]. 植物学报, 2022, 57(3): 288-298. |
| [9] | 李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41. |
| [10] | 刘栋. 既主内政, 又辖外交——以PHR为中心的基因网络调控植物-菌根真菌的共生[J]. 植物学报, 2021, 56(6): 647-650. |
| [11] | 李秋信, 迟伟, 季代丽. CURT1调控类囊体膜弯曲的研究进展[J]. 植物学报, 2021, 56(4): 462-469. |
| [12] | 车永梅, 孙艳君, 卢松冲, 侯丽霞, 范欣欣, 刘新. AtMYB77促进NO合成参与调控干旱胁迫下拟南芥侧根发育[J]. 植物学报, 2021, 56(4): 404-413. |
| [13] | 王婷, 羊欢欢, 赵弘巍, JosefVoglmeir, 刘丽. 蛋白质N-糖基化在拟南芥生长周期中的变化规律及去糖基化对根发育的影响[J]. 植物学报, 2021, 56(3): 262-274. |
| [14] | 林雨晴, 齐艳华. 生长素输出载体PIN家族研究进展[J]. 植物学报, 2021, 56(2): 151-165. |
| [15] | 马龙, 李桂林, 李师鹏, 蒋苏. 根尖整体透明技术改良[J]. 植物学报, 2020, 55(5): 596-604. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||