

植物学报 ›› 2021, Vol. 56 ›› Issue (6): 647-650.DOI: 10.11983/CBB21177 cstr: 32102.14.CBB21177
• 热点评述 • 下一篇
收稿日期:2021-10-12
接受日期:2021-10-18
出版日期:2021-11-01
发布日期:2021-11-12
通讯作者:
刘栋
作者简介:* E-mail: liu-d@mail.tsinghua.edu.cn基金资助:Received:2021-10-12
Accepted:2021-10-18
Online:2021-11-01
Published:2021-11-12
Contact:
Dong Liu
摘要: 磷是植物生长发育必需的大量矿质营养元素, 但自然界大部分土壤都存在严重缺磷的问题。为了适应这一营养逆境, 植物演化出一系列低磷胁迫应答反应。通过改变基因的转录水平调控低磷胁迫应答反应, 而转录因子PHR1在调控植物对低磷胁迫的转录响应中起关键作用。此外, 大部分陆生植物还能与丛枝菌根真菌建立共生关系, 通过丛枝菌根真菌更有效地从土壤中获取磷元素。最近, 中国科学院分子植物科学卓越创新中心王二涛研究组发现, 以PHR为中心的转录调控网络控制植物-丛枝菌根真菌共生的建立。因此, PHR不但在维持植物细胞自身的磷稳态中发挥作用, 而且参与植物与外界微生物的相互作用, 为植物有效地从环境中获得磷元素提供了另外一条途径。
刘栋. 既主内政, 又辖外交——以PHR为中心的基因网络调控植物-菌根真菌的共生. 植物学报, 2021, 56(6): 647-650.
Dong Liu. Managing Both Internal and Foreign Affairs—A PHR-centered Gene Network Regulates Plant-mycorrhizal Symbiosis. Chinese Bulletin of Botany, 2021, 56(6): 647-650.
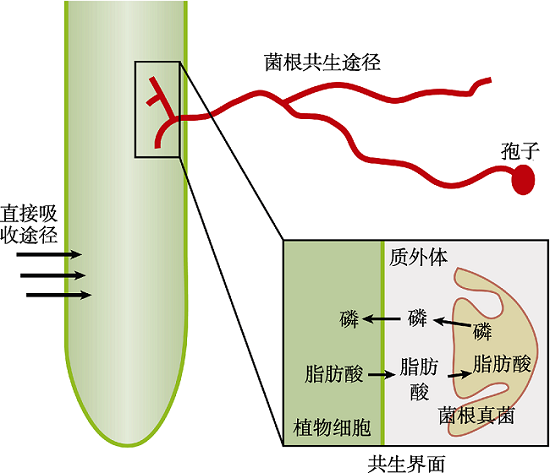
图1 植物从土壤中获取磷素的两种途径 (1) 根表皮细胞直接从土壤中吸收磷素; (2) 植物根皮层细胞与丛枝菌根真菌形成共生关系, 丛枝菌根真菌的菌丝从土壤中吸收磷素, 再提供给植物根组织。
Figure 1 Two pathways for plants to obtain Pi from soil (1) Root epidermal cells uptake Pi from soil directly; (2) Root cortex cells form symbiosis with arbuscular mycorrhiza fungi which uptake Pi from soil and provide them to root tissues.
| [1] | Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6, e1001102. |
| [2] |
Dong JS, Ma GJ, Sui LQ, Wei MW, Satheesh V, Zhang RY, Ge SH, Li JK, Zhang TE, Wittwer C, Jessen HJ, Zhang HM, An GY, Chao DY, Liu D, Lei MG (2019). Inositol pyrophosphate InsP8 acts as an intracellular phos- phate signal in Arabidopsis. Mol Plant 12, 1463-1473.
DOI URL |
| [3] |
Jiang YN, Wang WX, Xie QJ, Liu N, Liu LX, Wang DP, Zhang XW, Yang C, Chen XY, Tang DZ, Wang ET (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172-1175.
DOI PMID |
| [4] |
Liu F, Wang ZY, Ren HY, Shen CJ, Li Y, Ling HQ, Wu CY, Lian XM, Wu P (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62, 508-517.
DOI URL |
| [5] |
López-Arredondo DL, Leyva-González MA, González- Morales SI, López-Bucio J, Herrera-Estrella L (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65, 95-123.
DOI PMID |
| [6] |
Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plants. Science 356, 1175-1178.
DOI PMID |
| [7] |
Puga MI, Mateos I, Charukesi R, Wang ZY, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, Leyva A, Rubio V, Sommer H, Paz-Ares J (2014). SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111, 14947-14952.
DOI URL |
| [8] |
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15, 2122-2133.
DOI URL |
| [9] | Shi J, Zhao B, Zheng S, Zhang X, Wang X, Dong W, Xie Q, Gang W, Xiao Y, Chen F, Yu N, Wang E (2021). A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell https://doi.org/10.1016/j.cell.202109.030. |
| [10] |
Smith SE, Jakobsen I, Grønlund M, Smith FA (2011). Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol 156, 1050-1057.
DOI URL |
| [11] |
Sun LC, Song L, Zhang Y, Zheng Z, Liu D (2016). Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170, 499-514.
DOI URL |
| [12] |
Wang P, Snijders R, Kohlen W, Liu JY, Bisseling T, Limpens E (2021). Medicago SPX1 and SPX3 regulate phosphate homeostasis, mycorrhizal colonization, and arbuscule degradation. Plant Cell doi: 10.1093/plcell/koab206.
DOI |
| [13] |
Wang Z, Zheng Z, Song L, Liu D (2018). Functional charac- terization of Arabidopsis PHL4 in plant response to phosphate starvation. Front Plant Sci 9, 1432.
DOI PMID |
| [14] |
Wang ZY, Ruan WY, Shi J, Zhang L, Xiang D, Yang C, Li CY, Wu ZC, Liu Y, Yu YN, Shou HX, Mo XR, Mao CZ, Wu P (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111, 14953-14958.
DOI URL |
| [15] |
Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986-990.
DOI URL |
| [16] |
Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146, 1673-1686.
DOI PMID |
| [1] | 胡蝶, 蒋欣琪, 戴志聪, 陈戴一, 张雨, 祁珊珊, 杜道林. 丛枝菌根真菌提高入侵杂草南美蟛蜞菊对除草剂的耐受性[J]. 植物生态学报, 2024, 48(5): 651-659. |
| [2] | 陈科宇, 邢森, 唐玉, 孙佳慧, 任世杰, 张静, 纪宝明. 不同草地类型土壤丛枝菌根真菌群落特征及其驱动因素[J]. 植物生态学报, 2024, 48(5): 660-674. |
| [3] | 陈保冬, 付伟, 伍松林, 朱永官. 菌根真菌在陆地生态系统碳循环中的作用[J]. 植物生态学报, 2024, 48(1): 1-20. |
| [4] | 何斐, 李川, Faisal SHAH, 卢谢敏, 王莹, 王梦, 阮佳, 魏梦琳, 马星光, 王卓, 姜浩. 丛枝菌根菌丝桥介导刺槐-魔芋间碳转运和磷吸收[J]. 植物生态学报, 2023, 47(6): 782-791. |
| [5] | 杨佳绒, 戴冬, 陈俊芳, 吴宪, 刘啸林, 刘宇. 丛枝菌根真菌多样性对植物群落构建和稀有种维持的研究进展[J]. 植物生态学报, 2023, 47(6): 745-755. |
| [6] | 谢伟, 郝志鹏, 张莘, 陈保冬. 丛枝菌根网络介导的植物间信号交流研究进展及展望[J]. 植物生态学报, 2022, 46(5): 493-515. |
| [7] | 马炬峰, 辛敏, 徐陈超, 祝琬莹, 毛传澡, 陈欣, 程磊. 丛枝菌根真菌与氮添加对不同根形态基因型水稻氮吸收的影响[J]. 植物生态学报, 2021, 45(7): 728-737. |
| [8] | 庞芳, 夏维康, 何敏, 祁珊珊, 戴志聪, 杜道林. 固氮菌缓解氮限制环境中丛枝菌根真菌对加拿大一枝黄花的营养竞争[J]. 植物生态学报, 2020, 44(7): 782-790. |
| [9] | 韩雪, 苏锦权, 姚娜娜, 陈宝明. 外来入侵植物的根系觅养行为研究进展[J]. 生物多样性, 2020, 28(6): 727-733. |
| [10] | 崔利, 郭峰, 张佳蕾, 杨莎, 王建国, 孟静静, 耿耘, 李新国, 万书波. 摩西斗管囊霉改善连作花生根际土壤的微环境[J]. 植物生态学报, 2019, 43(8): 718-728. |
| [11] | 邹显花, 胡亚楠, 韦丹, 陈思同, 吴鹏飞, 马祥庆. 磷高效利用杉木对低磷胁迫的适应性与内源激素的相关性[J]. 植物生态学报, 2019, 43(2): 139-151. |
| [12] | 高文童, 张春艳, 董廷发, 胥晓. 丛枝菌根真菌对不同性别组合模式下青杨雌雄植株根系生长的影响[J]. 植物生态学报, 2019, 43(1): 37-45. |
| [13] | 徐丽娇, 郝志鹏, 谢伟, 李芳, 陈保冬. 丛枝菌根真菌根外菌丝跨膜H +和Ca 2+流对干旱胁迫的响应[J]. 植物生态学报, 2018, 42(7): 764-773. |
| [14] | 刘海跃, 李欣玫, 张琳琳, 王姣姣, 贺学礼. 西北荒漠带花棒根际丛枝菌根真菌生态地理分布[J]. 植物生态学报, 2018, 42(2): 252-260. |
| [15] | 陈思同, 邹显花, 蔡一冰, 韦丹, 李涛, 吴鹏飞, 马祥庆. 基于 32P示踪的不同供磷环境杉木幼苗磷的分配规律分析[J]. 植物生态学报, 2018, 42(11): 1103-1112. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
