

植物学报 ›› 2016, Vol. 51 ›› Issue (2): 175-183.DOI: 10.11983/CBB15054 cstr: 32102.14.CBB15054
收稿日期:2015-03-31
接受日期:2015-09-16
出版日期:2016-03-01
发布日期:2016-03-31
通讯作者:
E-mail: 基金资助:
Jiaxiao Du, Zongyan Qin, Si Xu, Xiang Jing, Ying Bao*( )
)
Received:2015-03-31
Accepted:2015-09-16
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: 摘要: 基于9个叶绿体基因片段(atpA、atpB、matK、petA、psaA、psbA、psbB、psbC和rbcL), 深入探讨了稻属(Oryza) 3个BBCC基因组异源四倍体和5个与之相关的BB或CC基因组二倍体物种间的谱系关系。进一步的系统发育分析表明: 3个具有相同BBCC基因组的四倍体物种并非同一次物种形成事件的产物, 而是在不同的分布区经历了至少3次分别的物种起源。其中, 四倍体Oryza punctata的母本可能来自同样分布在非洲并具有CC基因组的二倍体物种O. eichingeri; 而四倍体O. malampuzhaensis和O. minuta的母本则可能来自亚洲已经灭绝的具有BB基因组的不同二倍体。研究结果不但为追溯稻属异源四倍体的复杂网状进化提供了重要的分子证据, 而且拓展了我们对有花植物复杂物种形成的深入理解。
杜家潇, 秦宗燕, 徐思, 景翔, 包颖. 利用叶绿体基因探讨稻属BBCC基因组物种的系统起源. 植物学报, 2016, 51(2): 175-183.
Jiaxiao Du, Zongyan Qin, Si Xu, Xiang Jing, Ying Bao. Phylogenetic Origin of BBCC Genome Allotetraploids in Oryza Revealed by Chloroplast Gene Sequences. Chinese Bulletin of Botany, 2016, 51(2): 175-183.
| Species | Genome | IRRI Acc. No. | Origin |
|---|---|---|---|
| Oryza punctata Kotechy ex Steud. | BB | 103888 | Tanzania |
| 101434 | Tanzania | ||
| O. eichingeri A. Peter | CC | 105159 | Uganda |
| O. officinalis Wall. ex G. Watt. | CC | 104973 | China |
| O. rhizomatis D. A. Vaughan | CC | 103410 | Sri Lanka |
| O. malampuzhaensis Krish. et Chand. | BBCC | 80764 | India |
| 105223 | India | ||
| 105328 | India | ||
| O. minuta J. S. Presl. ex C. B. Presl. | BBCC | 104677 | Philippines |
| 105307 | Philippines | ||
| 103880 | Philippines | ||
| O. punctata | BBCC | 105181 | Uganda |
| 104059 | Nigeria | ||
| 104975 | Kenya | ||
| O. australiensis Domin | EE | 103303 | Australia |
| 105165 | Australia |
表1 稻属植物样品材料
Table 1 Plant materials of Oryza used in this study
| Species | Genome | IRRI Acc. No. | Origin |
|---|---|---|---|
| Oryza punctata Kotechy ex Steud. | BB | 103888 | Tanzania |
| 101434 | Tanzania | ||
| O. eichingeri A. Peter | CC | 105159 | Uganda |
| O. officinalis Wall. ex G. Watt. | CC | 104973 | China |
| O. rhizomatis D. A. Vaughan | CC | 103410 | Sri Lanka |
| O. malampuzhaensis Krish. et Chand. | BBCC | 80764 | India |
| 105223 | India | ||
| 105328 | India | ||
| O. minuta J. S. Presl. ex C. B. Presl. | BBCC | 104677 | Philippines |
| 105307 | Philippines | ||
| 103880 | Philippines | ||
| O. punctata | BBCC | 105181 | Uganda |
| 104059 | Nigeria | ||
| 104975 | Kenya | ||
| O. australiensis Domin | EE | 103303 | Australia |
| 105165 | Australia |
| Gene | Primer sequences (5'-3') |
|---|---|
| atpA | AGGCTTACTTGGGTCGTGTT TCCTCGGTGAATGTCTTGCT |
| atpB | ATCCTACTACTTCTCGTCCC AATTGCGATAATGTCCTGAA |
| petA | TTGGGTAAAGGAACAGATGA AATAACGGATGCGAAGAAGA |
| psaA | TAGCAGGGTTATTAGGACTTG ATGCGTGAATGTGATGGACT |
| psbA | TCGGATGGTTCGGTGTTTTGA AGGGAAGTTGTGAGCATTACG |
| psbB | GGGTTTGCCTTGGTATCGTG GGCTGTCTCCCTGTAGTTGG |
| psbC | GGGTTGATTTTACTTCCGCAC GAAGGTCCCAAAAACGCATAG |
| rbcL | TATCTTGGCAGCATTCCGAGTAA ATTTGATCGCTTTCCATATTTC |
表2 引物序列
Table 2 The sequences of primers
| Gene | Primer sequences (5'-3') |
|---|---|
| atpA | AGGCTTACTTGGGTCGTGTT TCCTCGGTGAATGTCTTGCT |
| atpB | ATCCTACTACTTCTCGTCCC AATTGCGATAATGTCCTGAA |
| petA | TTGGGTAAAGGAACAGATGA AATAACGGATGCGAAGAAGA |
| psaA | TAGCAGGGTTATTAGGACTTG ATGCGTGAATGTGATGGACT |
| psbA | TCGGATGGTTCGGTGTTTTGA AGGGAAGTTGTGAGCATTACG |
| psbB | GGGTTTGCCTTGGTATCGTG GGCTGTCTCCCTGTAGTTGG |
| psbC | GGGTTGATTTTACTTCCGCAC GAAGGTCCCAAAAACGCATAG |
| rbcL | TATCTTGGCAGCATTCCGAGTAA ATTTGATCGCTTTCCATATTTC |
| Gene name | Length (bp) | SNP | GC (%) |
|---|---|---|---|
| atpA | 997 | 3 | 42 |
| atpB | 673 | 3 | 43 |
| matK | 1552 | 16 | 34 |
| petA | 770 | 0 | 43 |
| psaA | 511 | 1 | 44 |
| psbA | 662 | 3 | 42 |
| psbB | 628 | 1 | 45 |
| psbC | 648 | 5 | 41 |
| rbcL | 1019 | 6 | 43 |
| Total | 7460 | 41 | 42 |
表3 9个稻属叶绿体基因片段的序列特点
Table 3 Sequence characteristics for 9 chloroplast gene fragments of Oryza
| Gene name | Length (bp) | SNP | GC (%) |
|---|---|---|---|
| atpA | 997 | 3 | 42 |
| atpB | 673 | 3 | 43 |
| matK | 1552 | 16 | 34 |
| petA | 770 | 0 | 43 |
| psaA | 511 | 1 | 44 |
| psbA | 662 | 3 | 42 |
| psbB | 628 | 1 | 45 |
| psbC | 648 | 5 | 41 |
| rbcL | 1019 | 6 | 43 |
| Total | 7460 | 41 | 42 |
| Species (genome) | Variable nucleotide sites | |||||||
|---|---|---|---|---|---|---|---|---|
| atpA | atpB | matK | psaA | psbA | psbB | psbC | rbcL | |
| O. punctata (BB) | GGA | GGC | CTATATAATTCTTTCT | C | ACC | G | GAAA | CTTGTC |
| O. punctata (BB) | ... | ... | ................ | . | ... | . | .... | ...... |
| O. minuta (BBCC) | A.. | ... | ................ | . | ... | . | .... | ...... |
| O. malampuzhaensis (BBCC) | A.. | A.. | .......C........ | . | ... | . | .... | ...... |
| O. eichingeri (CC) | AAC | AAT | TAGCGCCATCAACTTA | T | GTT | A | GGCG | ACGACG |
| O. officinalis (CC) | ... | ... | ........A....C.. | . | ... | . | C.A. | ...... |
| O. rhizomatis (CC) | ... | ... | ................ | . | ... | . | ..A. | ...... |
| O. punctata (BBCC) | ... | ... | ................ | . | ... | . | .... | ...... |
表4 9个稻属叶绿体基因片段的变异核苷酸位点
Table 4 Variable nucleotide sites of 9 chloroplast gene fragments of Oryza
| Species (genome) | Variable nucleotide sites | |||||||
|---|---|---|---|---|---|---|---|---|
| atpA | atpB | matK | psaA | psbA | psbB | psbC | rbcL | |
| O. punctata (BB) | GGA | GGC | CTATATAATTCTTTCT | C | ACC | G | GAAA | CTTGTC |
| O. punctata (BB) | ... | ... | ................ | . | ... | . | .... | ...... |
| O. minuta (BBCC) | A.. | ... | ................ | . | ... | . | .... | ...... |
| O. malampuzhaensis (BBCC) | A.. | A.. | .......C........ | . | ... | . | .... | ...... |
| O. eichingeri (CC) | AAC | AAT | TAGCGCCATCAACTTA | T | GTT | A | GGCG | ACGACG |
| O. officinalis (CC) | ... | ... | ........A....C.. | . | ... | . | C.A. | ...... |
| O. rhizomatis (CC) | ... | ... | ................ | . | ... | . | ..A. | ...... |
| O. punctata (BBCC) | ... | ... | ................ | . | ... | . | .... | ...... |
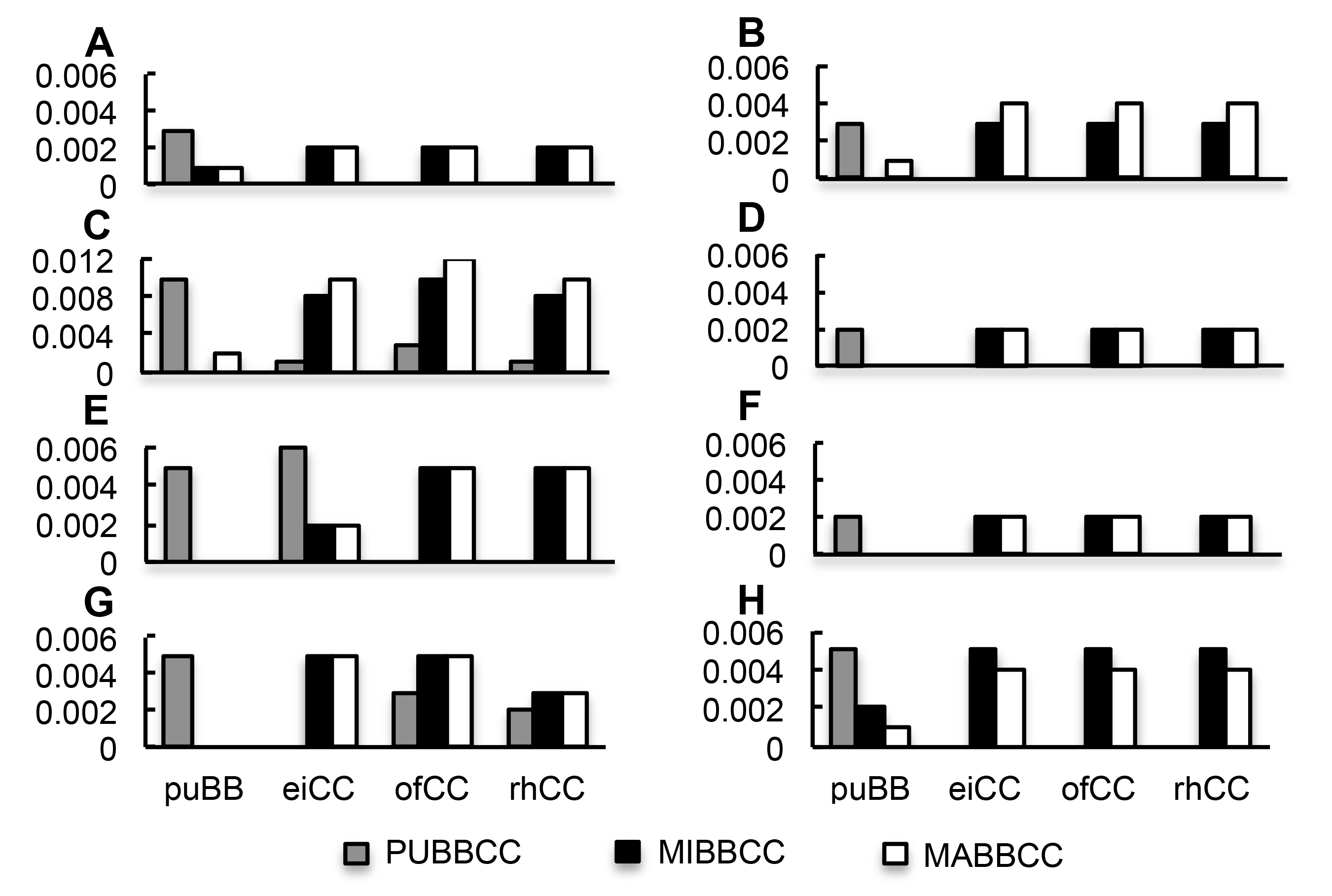
图1 BBCC基因组四倍体和其相关二倍体的对遗传距离 (A)-(H) 分别代表atpA、atpB、matK、psaA、psbA、psbB、psbC和rbcL基因片段。横坐标puBB、eiCC、ofCC和rhCC分别代表BB基因组的二倍体O. punctata和CC基因组的二倍体O. eichingeri、O. officinalis和O. rhizomatis; 纵坐标为遗传距离。
Figure 1 Pairwise genetic distances of the BBCC genomic tetraploids and their related diploids (A)-(H) Pairwise genetic distances of atpA, atpB, matK, psaA, psbA, psbB, psbC, and rbcL, respectively. x axis: puBB, eiCC, ofCC, and rhCC represents diploid O. punctata with genome BB, and O. eichingeri, O. officinalis, and O. rhizomatis with genome CC, respectively; y axis: Genetic distances

图2 叶绿体基因的最大似然树 (A)-(H) 分别基于个体基因atpA、atpB、matK、psaA、psbA、psbB、psbC和rbcL构建的最大似然性系统发育关系树; (I) 基于5个基因(atpA、atpB、matK、psbA和psbC)合并数据的最大似然树; 分支上的数字表示大于50%的自展支持率; BBCC基因组异源四倍体用粗体表示。
Figure 2 The maximum likelihood trees based on 8 and combined chloroplast genes (A)-(H) The ML tree based on atpA, atpB, matK, psaA, psbA, psbB, psbC, and rbcL gene, respectively; (I) The ML tree based on 5 combined genes (atpA, atpB, matK, psbA, and psbC). Numbers near branches indicate bootstrap values above 50%, Boldface indicates allotetraploid species with BBCC genome.
| [1] | Aggarwal RK, Brar DS, Khush GS (1997). Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254, 1-12. |
| [2] | Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999). Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98, 1320-1328. |
| [3] | Avise JC (2000). Phylogeography:the History and Forma- tion of Species. Cambridge: Harvard University Press. pp. 1-447. |
| [4] | Bao Y, Ge S (2003). Phylogenetic relationships among diploid species of Oryza officinalis complex revealed by multiple gene sequences. Acta Phytot Sin 41, 497-508. |
| [5] | Bao Y, Lu B, Ge S (2005). Identification of genomic consti- tutions of Oryza species with the B and C genomes by the PCR-RFLP method. Genet Res Crop Evol 52, 69-76. |
| [6] | Bao Y, Wendel FJ, Ge S (2010). Multiple patterns of rDNA evolution following polyploidy in Oryza. Mol Phylogenet Evol 55, 136-142. |
| [7] | Bao Y, Zhou HF, Ge S, Hong DY (2006). Genetic diversity and evolutionary relationships of Oryza species with the B- and C-genomes as revealed by SSR markers. J Plant Biol 49, 339-347. |
| [8] |
Buso GS, Rangel PH, Ferreira ME (2001). Analysis of random and specific sequences of nuclear and cytoplas- mic DNA in diploid and tetraploid American wild rice species (Oryza spp.). Genome 44, 476-494.
DOI PMID |
| [9] |
Criscuolo A (2011). morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol 61, 944-948.
DOI PMID |
| [10] |
Dally AM, Second G (1990). Chloroplast DNA diversity in wild and cultivated species of rice (genus Oryza, section Oryza). Cladistic-mutation and genetic-distance analysis. Theor Appl Genet 80, 209-222.
DOI PMID |
| [11] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [12] | Ge S, Li A, Lu BR, Zhang SZ, Hong DY (2002). A phylogeny of the rice tribe Oryzeae (Poaceae) based on matK sequence data. Am J Bot 89, 1967-1972. |
| [13] |
Ge S, Sang T, Lu BR, Hong DY (1999). Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96, 14400-14405.
DOI PMID |
| [14] |
Ge S, Sang T, Lu BR, Hong DY (2001). Rapid and reliable identification of rice genomes by RFLP analysis of PCR- amplified Adh genes. Genome 44, 1136-1142.
PMID |
| [15] |
Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27, 221-224.
DOI PMID |
| [16] |
Khush GS (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35, 25-34.
PMID |
| [17] | Kumagai M, Wang L, Ueda S (2010). Genetic diversity and evolutionary relationships in genus Oryza revealed by using highly variable regions of chloroplast DNA. Gene 462, 44-51. |
| [18] | Lu BR, Ge S, Sang T, Chen JK, Hong DY (2001). The current taxonomy and perplexity of the genus Oryza (Poaceae). Acta Phytot Sin 39, 373-388. |
| [19] |
Nishikawa T, Vaughan DA, Kadowaki K (2005). Phyloge- netic analysis of Oryza species, based on simple se- quence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theor Appl Genet 110, 696-705.
PMID |
| [20] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731-2739.
DOI PMID |
| [21] | Tian XJ, Zheng J, Hu SN, Yu J (2006). The rice mitochon- drial genomes and their variations. Plant Physiol 140, 401-410. |
| [22] | Vaughan DA (1994). The Wild Relatives of Rice:a Genetic Resources Handbook. Manila: International Rice Rese- arch Institute. pp. 1-137. |
| [23] | Wang BS, Ding ZY, Liu W, Pan J, Li CB, Ge S, Zhang DM (2009). Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol Biol 9, 250. |
| [24] |
Wing RA, Ammiraju JS, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, Brar D, Mackill DJ, Han B, Soderlund C, Stein L, SanMiguel P, Jackson S (2005). The Oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol 59, 53-62.
DOI PMID |
| [1] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 崔娟, 于晓玉, 于跃娇, 梁铖玮, 孙健, 陈温福. 影响中国东北和日本粳稻食味品质差异的质构因素及其遗传基础解析[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [5] | 张如礼, 李德铢, 张玉霄. 短穗竹居群遗传结构及气候适应性分析[J]. 植物学报, 2025, 60(3): 407-424. |
| [6] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [7] | 曹东, 李焕龙, 彭扬, 魏存争. 植物基因组大小与性状关系的研究进展[J]. 生物多样性, 2025, 33(2): 24192-. |
| [8] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [9] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [10] | 姚祥坦, 张心怡, 陈阳, 袁晔, 程旺大, 王天瑞, 邱英雄. 基于基因组重测序揭示栽培欧菱遗传多样性及‘南湖菱’的起源驯化历史[J]. 生物多样性, 2024, 32(9): 24212-. |
| [11] | 张强, 赵振宇, 李平华. 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024, 59(6): 978-998. |
| [12] | 李园, 范开建, 安泰, 李聪, 蒋俊霞, 牛皓, 曾伟伟, 衡燕芳, 李虎, 付俊杰, 李慧慧, 黎亮. 玉米自然群体自交系农艺性状的多环境全基因组预测初探[J]. 植物学报, 2024, 59(6): 1041-1053. |
| [13] | 陈文娜, 李良涛, 周璐, 姚纲. 太行山近期隆升促进太行花属(蔷薇科)谱系分化[J]. 植物学报, 2024, 59(5): 763-773. |
| [14] | 胡丹玲, 孙永伟. 病毒介导的植物基因组编辑技术研究进展[J]. 植物学报, 2024, 59(3): 452-462. |
| [15] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||