

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (3): 373-384.DOI: 10.11983/CBB22063 cstr: 32102.14.CBB22063
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Shuyao Zhou, Jianming Li, Juan Mao( )
)
Received:2022-04-05
Accepted:2022-07-25
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: maojuan@scau.edu.cn
Shuyao Zhou, Jianming Li, Juan Mao. AtGH3.17-mediated Regulation of Auxin and Brassinosteroid Response in Arabidopsis thaliana[J]. Chinese Bulletin of Botany, 2023, 58(3): 373-384.
| Primer name | Primer sequence (5′-3′) |
|---|---|
| pBRI1:gGH3.17-F | CAAACTCTTGAGAAGGTACCATGATA- CCAAGTTACGACCC |
| pBRI1:gGH3.17-R | GTGTCGACTCTAGAGGATCCAGAAT- CTAAACCAAGTGGTT |
| GH3.17-DT1-BsF | ATATATGGTCTCGATTGGATCTCATGTCCTAAGAGTGTTTTAGAGCTAGAAATAGC |
| GH3.17-DT1-BsR | ATTATTGGTCTCGAAACGCAGAGACTCGGTCTTGTCCAATCTCTTAGTCGACTCTAC |
| U6-26p-F | TGTCCCAGGATTAGAATGATTAGGC |
| U6-26p-R | AGCCCTCTTCTTTCGATCCATCAAC |
| CAS 9-F | ATCCAATCTTCGGCAACAT |
| CAS 9-R | TATCCAGGTCATCGTCGTA |
| GH3.17-ba-F | AGATGAGGGAAAAGGGATGT |
| GH3.17-ba-R | TCAAGATTCCTTCCCACGAC |
| GH3.17-qPCR-F | CGATGTATGCTTCCTCTGAGTG |
| GH3.17-qPCR-R | ATCTCTTCGTGGGATTTGTCG |
| IAA12-qPCR-F | TGGGTTACACAGGATGAACAG |
| IAA12-qPCR-R | AACCCTAAGCCCTGAACTTTC |
| IAA16-qPCR-F | TGAAGATAAAGATGGCGACTGG |
| IAA16-qPCR-R | AAGTCCGATTGCTTCTGATCC |
| IAA20-qPCR-F | GTACTCGAAACCTAAGCACGG |
| IAA20-qPCR-R | CACATATTCCGCATCCTCTACC |
| DWF4-qPCR-F | CATTGCTCTCGCTATCTTCTTC |
| DWF4-qPCR-R | GACTCTCCTAGTTCCTTCTTGG |
| CPD-qPCR-F | TCCTTGTGGGTCTAGTGTTTG |
| CPD-qPCR-R | TTGAACCATTGAAGCAGAAGAG |
| BES1-qPCR-F | CAGCCATTCTCTGCCTCTATG |
| BES1-qPCR-R | ACTCGGAGCTTTGACCAATC |
| Actin2-qPCR-F | GGTAACATTGTGCTCAGTGGTGG |
| Actin2-qPCR-R | AACGACCTTAATCTTCATGCTGC |
Table 1 Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| pBRI1:gGH3.17-F | CAAACTCTTGAGAAGGTACCATGATA- CCAAGTTACGACCC |
| pBRI1:gGH3.17-R | GTGTCGACTCTAGAGGATCCAGAAT- CTAAACCAAGTGGTT |
| GH3.17-DT1-BsF | ATATATGGTCTCGATTGGATCTCATGTCCTAAGAGTGTTTTAGAGCTAGAAATAGC |
| GH3.17-DT1-BsR | ATTATTGGTCTCGAAACGCAGAGACTCGGTCTTGTCCAATCTCTTAGTCGACTCTAC |
| U6-26p-F | TGTCCCAGGATTAGAATGATTAGGC |
| U6-26p-R | AGCCCTCTTCTTTCGATCCATCAAC |
| CAS 9-F | ATCCAATCTTCGGCAACAT |
| CAS 9-R | TATCCAGGTCATCGTCGTA |
| GH3.17-ba-F | AGATGAGGGAAAAGGGATGT |
| GH3.17-ba-R | TCAAGATTCCTTCCCACGAC |
| GH3.17-qPCR-F | CGATGTATGCTTCCTCTGAGTG |
| GH3.17-qPCR-R | ATCTCTTCGTGGGATTTGTCG |
| IAA12-qPCR-F | TGGGTTACACAGGATGAACAG |
| IAA12-qPCR-R | AACCCTAAGCCCTGAACTTTC |
| IAA16-qPCR-F | TGAAGATAAAGATGGCGACTGG |
| IAA16-qPCR-R | AAGTCCGATTGCTTCTGATCC |
| IAA20-qPCR-F | GTACTCGAAACCTAAGCACGG |
| IAA20-qPCR-R | CACATATTCCGCATCCTCTACC |
| DWF4-qPCR-F | CATTGCTCTCGCTATCTTCTTC |
| DWF4-qPCR-R | GACTCTCCTAGTTCCTTCTTGG |
| CPD-qPCR-F | TCCTTGTGGGTCTAGTGTTTG |
| CPD-qPCR-R | TTGAACCATTGAAGCAGAAGAG |
| BES1-qPCR-F | CAGCCATTCTCTGCCTCTATG |
| BES1-qPCR-R | ACTCGGAGCTTTGACCAATC |
| Actin2-qPCR-F | GGTAACATTGTGCTCAGTGGTGG |
| Actin2-qPCR-R | AACGACCTTAATCTTCATGCTGC |
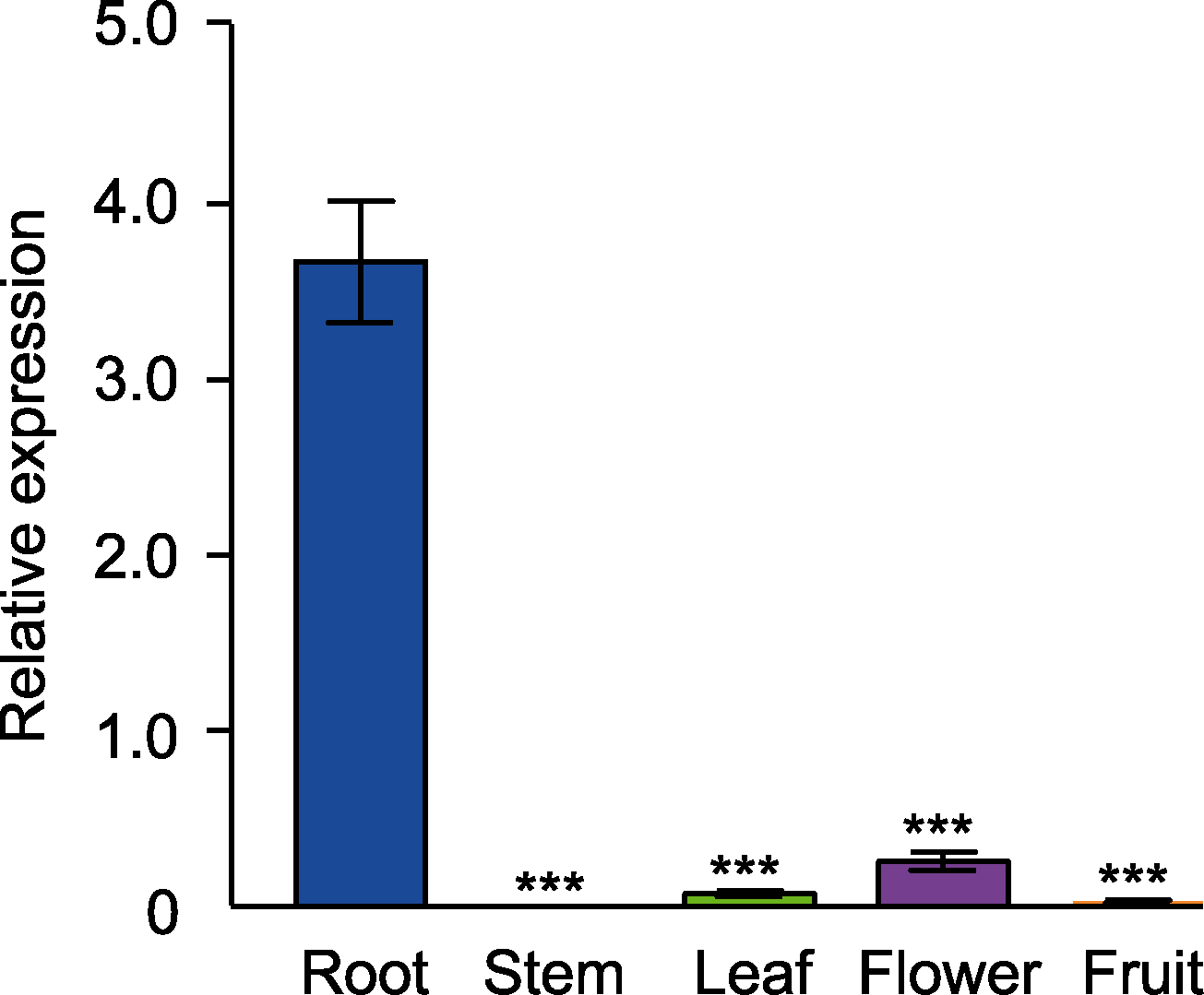
Figure 1 Tissue-specific expression of GH3.17 in Arabidopsis thaliana Error bars denote SEM. *** indicate significant differences at P<0.001 (Student’s t-test).
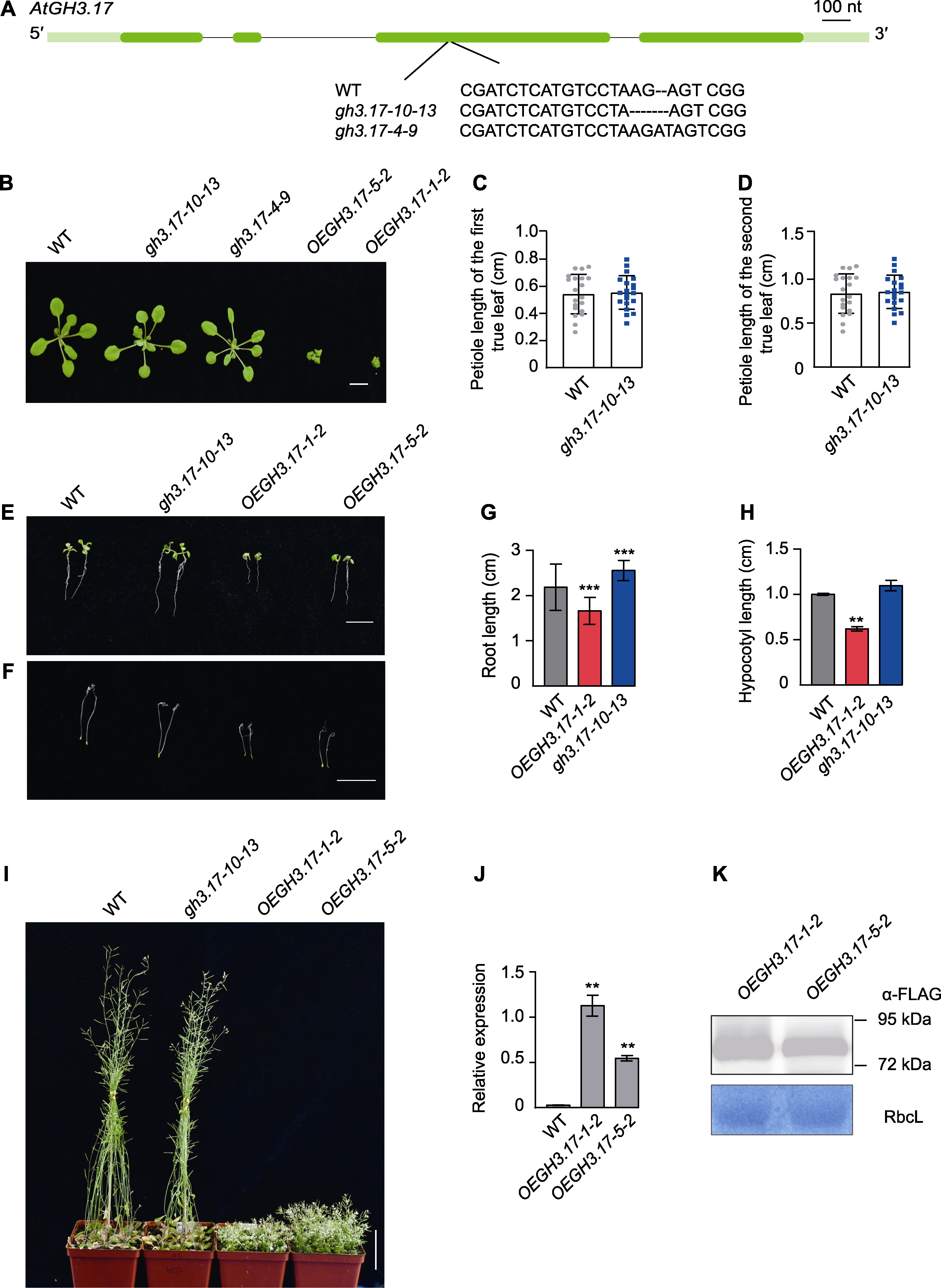
Figure 2 Phenotype of Arabidopsis thaliana wild type (WT), GH3.17 CRISPR mutant and overexpression seedlings (A) Generation of knockout alleles of GH3.17 (gh3.17-10-13 and gh3.17-4-9) by CRISPR/Cas9 (exons are represented by dark green boxes, introns by black lines and 5' and 3' untranslated region (UTR) by light green boxes; the mutation site and sequence are presented in the figure); (B) The aerial part phenotype of 20-day-old soil-grown WT, GH3.17 CRISPR mutant and overexpression lines under normal conditions (bar=1 cm); (C) Petiole length statistics of the first pair of true leaves of WT and GH3.17 CRISPR mutant (n=20); (D) Statistics of petiole length of the second pair of true leaves of WT and GH3.17 CRISPR mutant (n=20); (E), (G) The primary root length phenotype and statistics of 7-day-old WT, GH3.17 CRISPR mutant and overexpression seedlings vertically grown on 1/2MS medium under normal conditions (bar=1 cm) (the data was obtained through three replicates of 30 seedlings each); (F), (H) The hypocotyls phenotype and statistics of 5-day-old dark-grown WT, GH3.17 CRISPR mutant and overexpression seedlings grown on 1/2MS medium under normal conditions (bar=1 cm) (the data was obtained through three replicates of 30 seedlings each); (I) The above-ground phenotypes of 50-day-old soil-grown WT, GH3.17 CRISPR mutant and overexpression lines under normal conditions (bar=5 cm); (J) qRT-PCR analysis of GH3.17 transcriptional level in WT and GH3.17 overexpression plants; (K) Immunoblot analysis of GH3.17-Flag fusion protein in GH3.17 overexpression transgenic lines. Error bars in (C), (D), (G) and (H) denote SD; error bars in (J) denote SEM. ** indicate significant differences at P<0.01; *** indicate significant differences at P<0.001 (Student’s t-test).
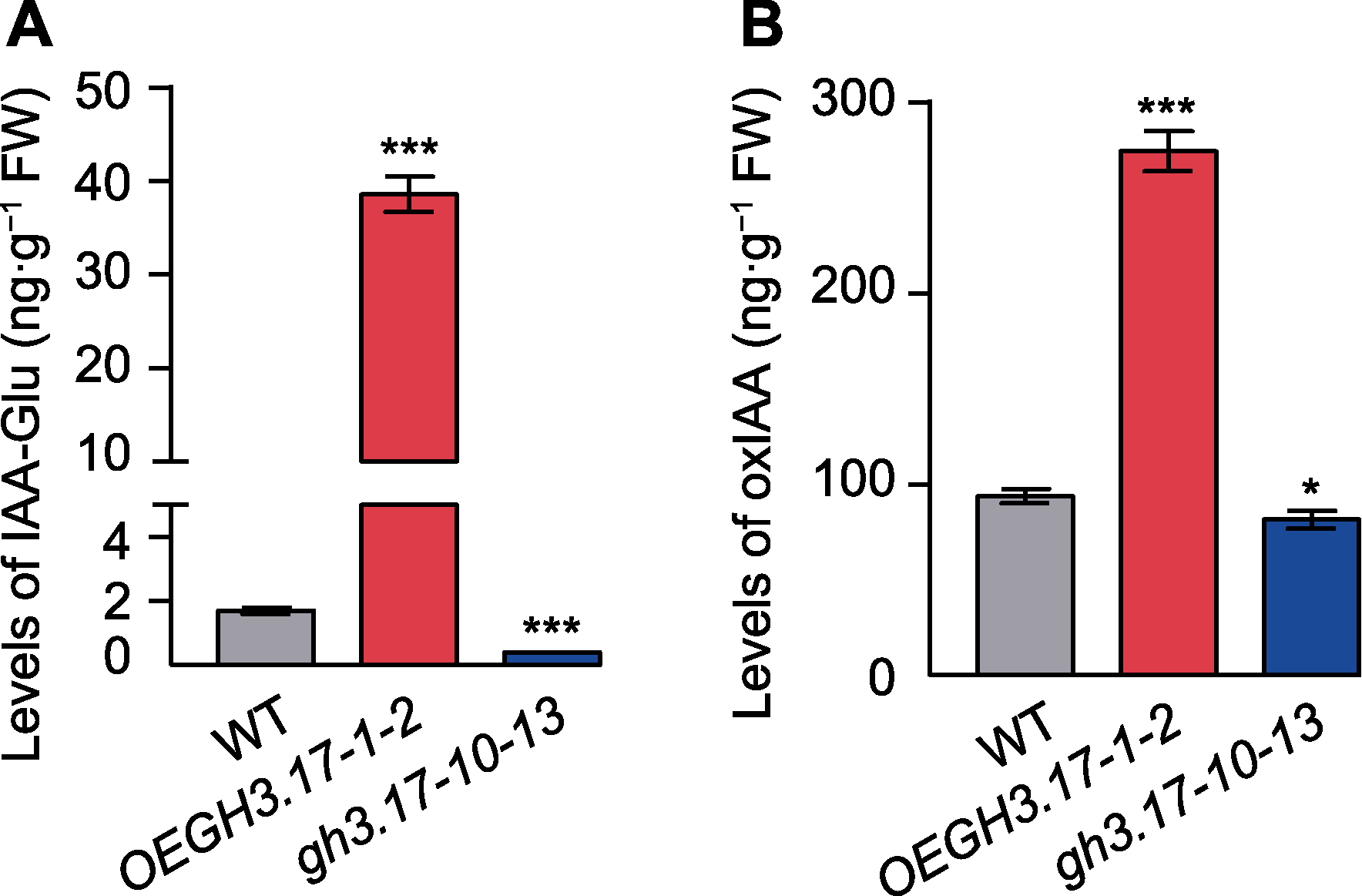
Figure 3 The IAA-Glu and oxIAA content of Arabidopsis tha- liana wild type (WT), GH3.17 overexpression and CRISPR mutant (A) The IAA-Glu content of WT, GH3.17 overexpression and CRISPR mutant; (B) The oxIAA content of WT, GH3.17 overexpression and CRISPR mutant. IAA level in 12-day-old Arabidopsis thaliana seedlings grown on 1/2MS solid medium is obtained from extraction of three independent tissue samples. Error bars indicate SD. * indicates significant difference at P<0.05; *** indicate significant differences at P<0.001 (Student’s t-test).
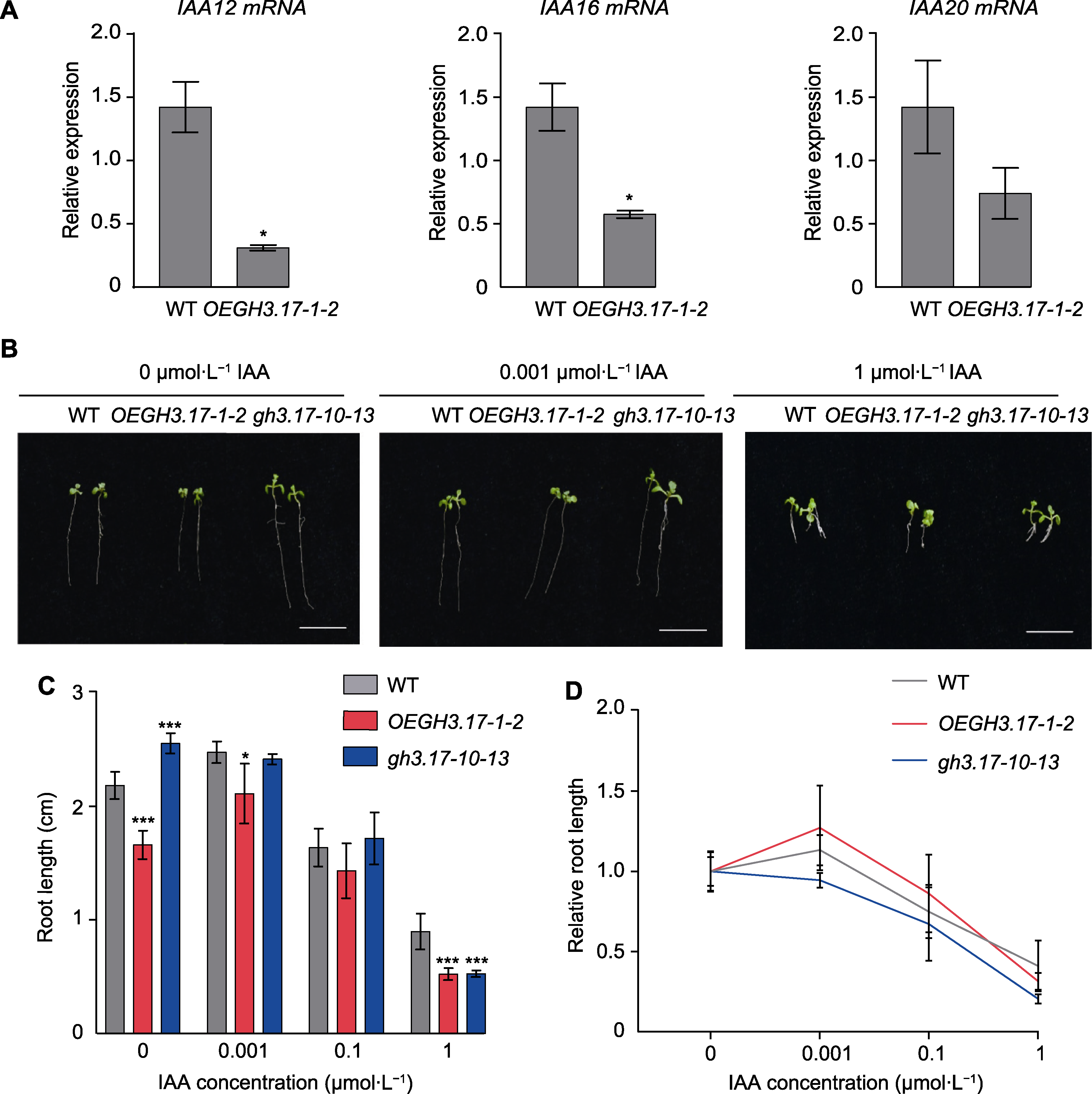
Figure 4 Response of Arabidopsis thaliana wild type (WT), GH3.17 CRISPR mutant and overexpression seedlings to auxin (A) The relative expression level of IAA12, IAA16 and IAA20 in WT and OEGH3.17-1-2 overexpression plants (error bars represent SEM of three technical duplicates); (B) The root phenotype of WT, GH3.17 CRISPR mutant and overexpression seedling on 1/2MS medium with different concentrations of IAA for 7 days (bars=1 cm); (C) The statistical analysis of WT, GH3.17 CRISPR mutant and overexpression seedling root length grown on 1/2MS medium with different concentrations of IAA for 7 days (the data was obtained through three replicates of 30 seedlings each, error bars represent SD); (D) Root length curves of WT, GH3.17 CRISPR mutant and overexpression seedling showing their response to different concentrations of IAA (the data was obtained through three replicates of 30 seedlings each, error bars represent SD). * indicate significant differences at P<0.05; *** indicate significant differences at P<0.001 (Student’s t-test).
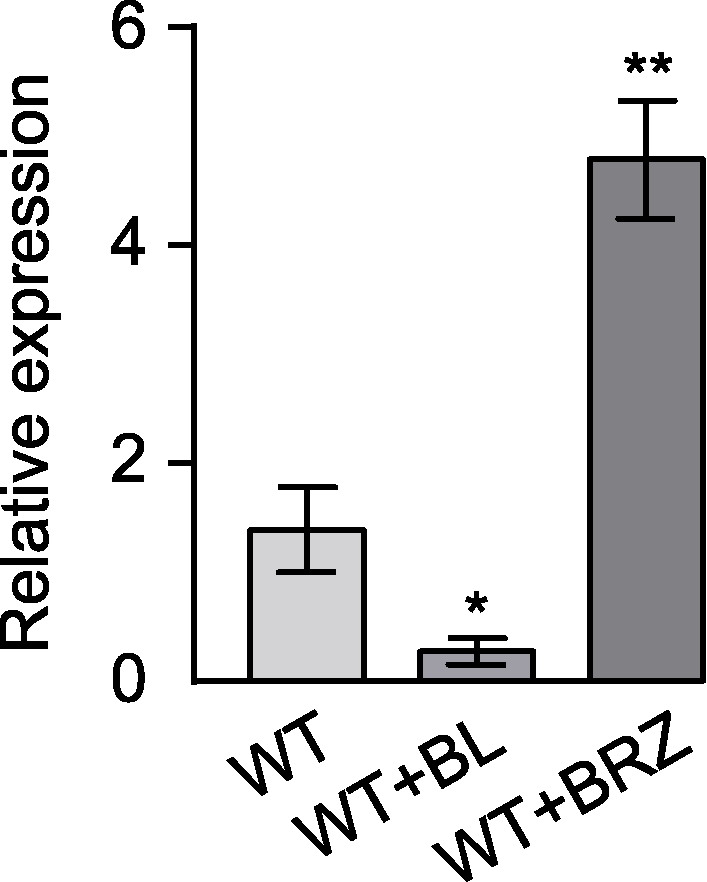
Figure 5 The transcription level of Arabidopsis thaliana GH3.17 was inhibited by brassinolide (BL) treatment Error bars represent SEM of three technical duplicates. * indicates significant difference at P<0.05; ** indicates significant difference at P<0.01 (Student’s t-test). WT: Wild type
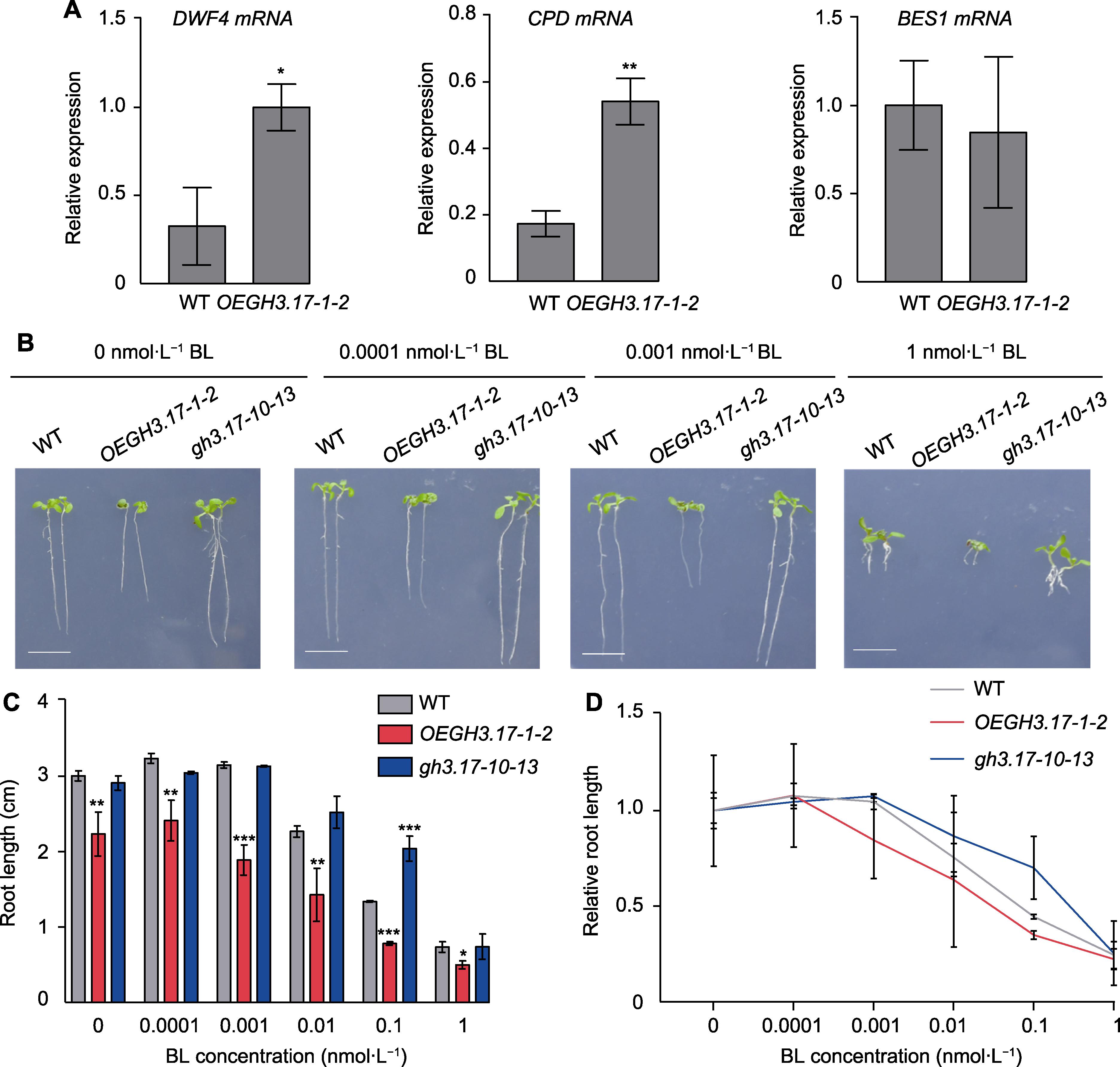
Figure 6 Response of Arabidopsis thaliana wild type (WT), GH3.17 overexpression and CRISPR mutant seedlings to brassinosteroid (BR) (A) The relative expression level of DWF4, CPD and BES1 in WT and OEGH3.17-1-2 overexpression plants by qRT-PCR (error bars represent SEM of three technical duplicates); (B) The root phenotype of WT, OEGH3.17-1-2 and gh3.17-10-13 grown on 1/2MS solid medium with different concentrations of brassinolide (BL) for 9 days (bars=1 cm); (C) The statistical analysis of WT, OEGH3.17-1-2 and gh3.17-10-13 root length grown on 1/2MS solid medium with different concentrations of BL for 9 days (the data was obtained through three replicates of 30 seedlings each, error bars represent SD); (D) Root length curves of WT, OEGH3.17-1-2 and gh3.17-10-13 showing their response to different concentrations of BL (the data was obtained through three replicates of 30 seedlings each, error bars represent SD). * indicate significant differences at P<0.05; ** indicate significant differences at P<0.01; *** indicate significant differences at P<0.001 (Student’s t-test).
| [1] |
李艳艳, 齐艳华 (2022). 植物Aux/IAA基因家族生物学功能研究进展. 植物学报 57, 30-41.
DOI |
| [2] |
任鸿雁, 王莉, 马青秀, 吴光 (2015). 油菜素内酯生物合成途径的研究进展. 植物学报 50, 768-778.
DOI |
| [3] | 孙超, 黎家 (2017). 油菜素甾醇类激素的生物合成、代谢及信号转导. 植物生理学报 53, 291-307. |
| [4] | 王冰, 李家洋, 王永红 (2006). 生长素调控植物株型形成的研究进展. 植物学通报 23, 443-458. |
| [5] |
谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光 (2021). 基于CRISPR编辑系统的DNA片段删除技术. 植物学报 56, 44-49.
DOI |
| [6] |
Abel S, Theologis A (1996). Early genes and auxin action. Plant Physiol 111, 9-17.
DOI PMID |
| [7] |
Bao F, Shen JJ, Brady SR, Muday GK, Asami T, Yang ZB (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134, 1624-1631.
DOI URL |
| [8] |
Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, Park T, Cho H, Hwang I, Choe S (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66, 564-578.
DOI URL |
| [9] |
Clouse SD, Sasse JM (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49, 427-451.
DOI URL |
| [10] | Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Marée AFM, Costantino P, Grieneisen VA, Sabatini S (2017). Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 114, E7641-E7649. |
| [11] |
Di Mambro R, Svolacchia N, Dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, Busch W, Costantino P, Sabatini S (2019). The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol 29, 1199-1205.
DOI PMID |
| [12] |
Ding ZJ, Friml J (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107, 12046-12051.
DOI URL |
| [13] | Du MM, Spalding EP, Gray WM (2020). Rapid auxin-mediated cell expansion. Annu Rev Plant Biol 71, 379-402. |
| [14] |
Favero DS, Le KN, Neff MM (2017). Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J 89, 1133-1145.
DOI URL |
| [15] |
Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134, 1555-1573.
DOI URL |
| [16] |
Hagen G, Guilfoyle T (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49, 373-385.
PMID |
| [17] |
Hagen G, Guilfoyle TJ (1985). Rapid induction of selective transcription by auxins. Mol Cell Biol 5, 1197-1203.
DOI PMID |
| [18] |
Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2017). SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol Plant 10, 590-604.
DOI PMID |
| [19] |
Hsieh HL, Okamoto H, Wang ML, Ang LH, Matsui M, Goodman H, Deng XW (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14, 1958-1970.
DOI URL |
| [20] |
Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI (2009). Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA 106, 13630-13635.
DOI PMID |
| [21] |
Liu XL, Yang HX, Wang Y, Zhu ZH, Zhang W, Li JM (2020). Comparative transcriptomic analysis to identify b- rassinosteroid response genes. Plant Physiol 184, 1072-1082.
DOI URL |
| [22] |
Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994). Soybean GH3promoter contains multiple auxin-inducible elements. Plant Cell 6, 645-657.
PMID |
| [23] |
Luo J, Zhou JJ, Zhang JZ (2018). Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci 19, 259.
DOI URL |
| [24] |
Mouchel CF, Osmont KS, Hardtke CS (2006). BRX mediates feedback between brassinosteroid levels and auxin signaling in root growth. Nature 443, 458-461.
DOI |
| [25] |
Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133, 1843-1853.
PMID |
| [26] |
Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25, 213-221.
PMID |
| [27] |
Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, Szekeres M, Mizutani M (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18, 3275-3288.
DOI URL |
| [28] |
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu GX, Theologis A (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444-463.
DOI PMID |
| [29] |
Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106, 22540-22545.
DOI PMID |
| [30] |
Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73, 676-688.
DOI URL |
| [31] |
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616-627.
DOI PMID |
| [32] |
Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A (1994). Biosynthesis of brassinolide from teasterone via typhasterol and castasterone in cultured cells of Catharanthus roseus. J Plant Growth Regul 13, 21-26.
DOI URL |
| [33] |
Tan X, Calderon-Villalobos LIA, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645.
DOI |
| [34] |
Ulmasov T, Hagen G, Guilfoyle TJ (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865-1868.
DOI URL |
| [35] |
Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995). Composite structure of auxin response elements. Plant Cell 7, 1611-1623.
PMID |
| [36] | Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963-1971. |
| [37] |
Wang ZY, Bai MY, Oh E, Zhu JY (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46, 701-724.
DOI URL |
| [38] |
Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24, 1874-1885.
DOI PMID |
| [39] |
Woodward AW, Bartel B (2005). Auxin: regulation, action, and interaction. Ann Bot 95, 707-735.
DOI URL |
| [40] | Yoshimitsu Y, Tanaka K, Fukuda W, Asami T, Yoshida S, Hayashi KI, Kamiya Y, Jikumaru Y, Shigeta T, Nakamura Y, Matsuo T, Okamoto S (2011). Transcription of DWARF4 plays a crucial role in auxin-regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PLoS One 6, e23851. |
| [41] |
Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X, Chen Y, Chen Q (2018). Evolutionary history of the Glycoside Hydrolase 3 (GH3) family based on the sequenced genomes of 48 plants and identification of jasmonic acid- related GH3 proteins in Solanum tuberosum. Int J Mol Sci 19, 1850.
DOI URL |
| [42] |
Zheng ZY, Guo YX, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016). Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2, 16025.
DOI PMID |
| [1] | Yuying Zhou, Hui Chen, Simu Liu. Research Progress on Auxin Responsive Non-canonical Aux/IAA Proteins in Plants [J]. Chinese Bulletin of Botany, 2024, 59(4): 651-658. |
| [2] | Xiangpei Kong, Mengyue Zhang, Zhaojun Ding. There Is a Way Out-new Breakthroughs in Extracellular Auxin Sensing [J]. Chinese Bulletin of Botany, 2023, 58(6): 861-865. |
| [3] | Yuan Yuan, Enhebayaer, Qi Yanhua. Research Advances in Biological Functions of GH3 Gene Family in Plants [J]. Chinese Bulletin of Botany, 2023, 58(5): 770-782. |
| [4] | Ming-Yi Bai, Jinrong Peng, Xiangdong Fu. Coordinated Regulation of Gibberellin and Brassinosteroid Signalings Drives Toward a Sustainable “Green Revolution” by Breeding the New Generation of High-yield Wheat [J]. Chinese Bulletin of Botany, 2023, 58(2): 194-198. |
| [5] | Ye Qing, Yan Xiaoyan, Chen Huize, Feng Jinlin, Han Rong. Effect of Nitrogen-doped Graphene Quantum Dots on Growth Direction of Primary Root in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2022, 57(5): 623-634. |
| [6] | Lixia Jia, Yanhua Qi. Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction [J]. Chinese Bulletin of Botany, 2022, 57(3): 263-275. |
| [7] | Binqi Li, Jiahui Yan, Hao Li, Wei Xin, Yunhe Tian, Zhenbiao Yang, Wenxin Tang. Changes of Small GTPases Activity During Cucumber Tendril Winding [J]. Chinese Bulletin of Botany, 2022, 57(3): 299-307. |
| [8] | Jingwen Wang, Xingjun Wang, Changle Ma, Pengcheng Li. A Review on the Mechanism of Ribosome Stress Response in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 80-89. |
| [9] | Yanyan Li, Yanhua Qi. Advances in Biological Functions of Aux/IAA Gene Family in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 30-41. |
| [10] | Yuqing Lin, Yanhua Qi. Advances in Auxin Efflux Carrier PIN Proteins [J]. Chinese Bulletin of Botany, 2021, 56(2): 151-165. |
| [11] | Rongfeng Huang, Tongda Xu. Auxin Regulates the Lateral Root Development Through MAPK-mediated VLCFAs Biosynthesis [J]. Chinese Bulletin of Botany, 2021, 56(1): 6-9. |
| [12] | Yuting Yao,Jiaqi Ma,Xiaoli Feng,Jianwei Pan,Chao Wang. A Role of Arabidopsis Phosphoinositide Kinase, FAB1, in Root Hair Growth [J]. Chinese Bulletin of Botany, 2020, 55(2): 126-136. |
| [13] | Shuhui Zhang,Hong Wang,Wenru Wang,Xuelian Wu,Yuansong Xiao,Futian Peng. Effects of Sucrose on Seedling Growth and Development and SnRK1 Activity in Prunus persica [J]. Chinese Bulletin of Botany, 2019, 54(6): 744-752. |
| [14] | Zhenmei He,Dongming Li,Yanhua Qi. Advances in Biofunctions of the ABCB Subfamily in Plants [J]. Chinese Bulletin of Botany, 2019, 54(6): 688-698. |
| [15] | Kongqin Hu, Zhaojun Ding. A TIR1-independent Auxin Signaling Module [J]. Chinese Bulletin of Botany, 2019, 54(3): 293-295. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||