

Chinese Bulletin of Botany ›› 2025, Vol. 60 ›› Issue (5): 749-758.DOI: 10.11983/CBB25106 cstr: 32102.14.CBB25106
• INVITED REVIEWS • Previous Articles Next Articles
Li Wenliang1, Feng Hanqing1,*( ), Lai Jianbin2,*(
), Lai Jianbin2,*( )
)
Received:2025-06-06
Accepted:2025-07-01
Online:2025-09-10
Published:2025-07-02
Contact:
Feng Hanqing, Lai Jianbin
Li Wenliang, Feng Hanqing, Lai Jianbin. Research Progress on SUMOylation in Plant-Pathogen Interactions[J]. Chinese Bulletin of Botany, 2025, 60(5): 749-758.
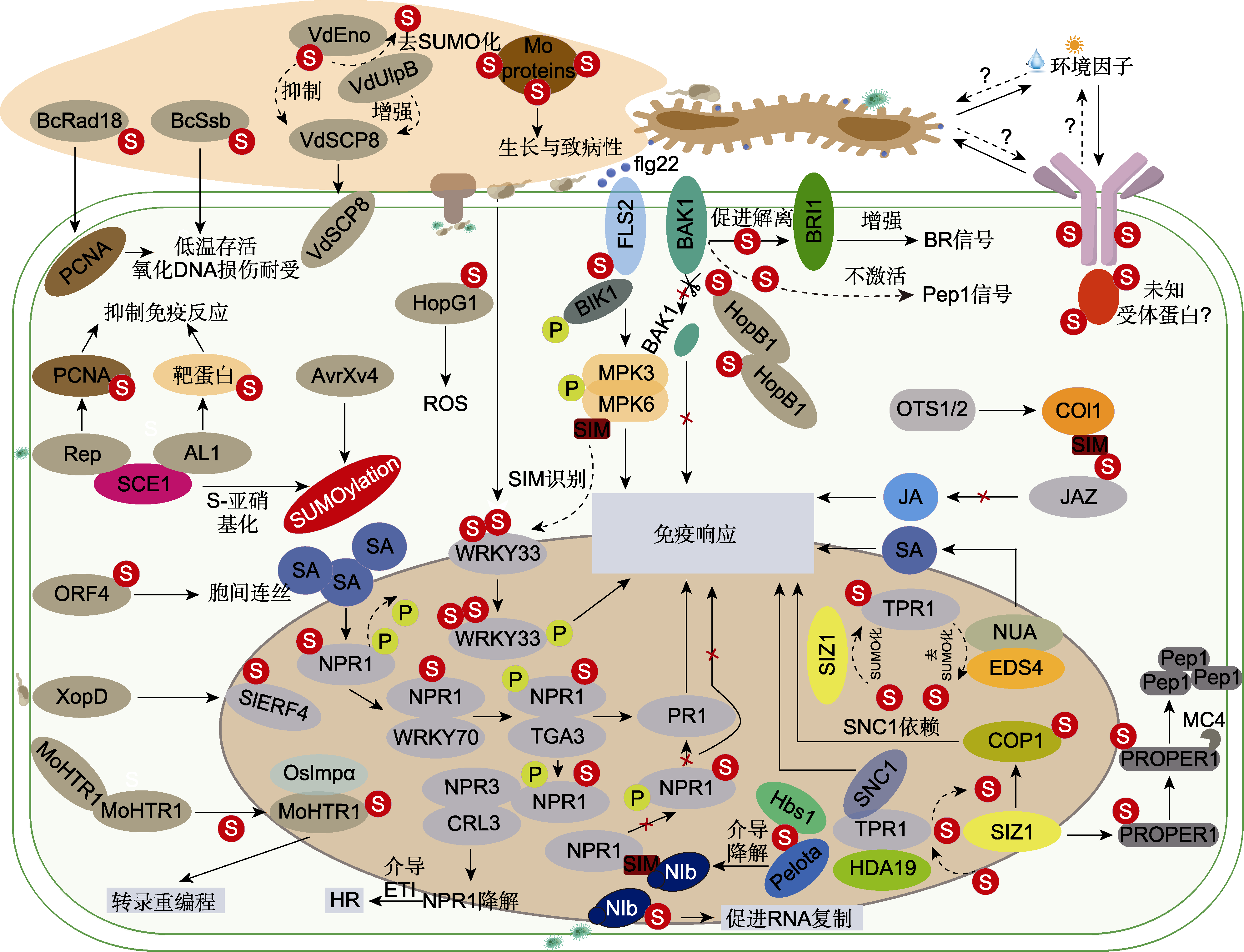
Figure 1 The immuneity regulatory networks of SUMOylation in plant-pathogen interactions During plant-pathogen interactions, SUMOylation regulates the immune functions of fungal pathogenicity proteins, effector proteins and immunity-associated proteins in plant cells, as well as key transcription factors. S: SUMOylation; P: Phosphorylation; Mo: Magnaporthe oryzae (rice blast fungus); Bc: Botrytis cinerea (gray mold fungus); Vd: Verticillium dahliae (verticillium wilt fungus); SA: Salicylic acid; JA: Jasmonic acid; HR: Hypersensitive response; ROS: Reactive oxygen species; BR: Brassinosteroids; ETI: Effector-triggered immunity; X: Inhibition. Solid lines indicate direct targeting regulatory effects; Dash lines indicate the bidirec tional nature of molecular regulation.
| [1] | An YY, Zhang MX (2024). Advances in understanding the plant-Ralstonia solanacearum interactions: unraveling the dynamics, mechanisms, and implications for crop disease resistance. New Crops 1, 100014. |
| [2] | Arroyo-Mateos M, Sabarit B, Maio F, Sánchez-Durán MA, Rosas-Díaz T, Prins M, Ruiz-Albert J, Luna AP, van den Burg HA, Bejarano ER (2018). Geminivirus replication protein impairs SUMO conjugation of proliferating cellular nuclear antigen at two acceptor sites. J Virol 92, e00611-18. |
| [3] |
Augustine RC, York SL, Rytz TC, Vierstra RD (2016). Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol 171, 2191-2210.
DOI PMID |
| [4] |
Castaño-Miquel L, Mas A, Teixeira I, Seguí J, Perearnau A, Thampi BN, Schapire AL, Rodrigo N, La Verde G, Manrique S, Coca M, Lois LM (2017). SUMOylation inhibition mediated by disruption of SUMO E1-E2 interactions confers plant susceptibility to necrotrophic fungal pathogens. Mol Plant 10, 709-720.
DOI PMID |
| [5] |
Castaño-Miquel L, Seguí J, Manrique S, Teixeira I, Carretero-Paulet L, Atencio F, Lois LM (2013). Diversification of SUMO-activating enzyme in Arabidopsis: implications in SUMO conjugation. Mol Plant 6, 1646-1660.
DOI PMID |
| [6] | Cheng XF, Xiong RY, Li YZ, Li FF, Zhou XP, Wang AM (2017). SUMOylation of Turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 29, 508-525. |
| [7] |
Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol 151, 1930-1942.
DOI PMID |
| [8] | Colignon B, Dieu M, Demazy C, Delaive E, Muhovski Y, Raes M, Mauro S (2017). Proteomic study of SUMOylation during Solanum tuberosum-Phytophthora infestans interactions. Mol Plant Microbe Interact 30, 855-865. |
| [9] | Fu ZQ, Yan SP, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, Dong XN (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228-232. |
| [10] | Ge LH, Cao BW, Qiao R, Cui HG, Li SF, Shan HY, Gong P, Zhang MZ, Li H, Wang AM, Zhou XP, Li FF (2023). SUMOylation-modified Pelota-Hbs1 RNA surveillance com- plex restricts the infection of potyvirids in plants. Mol Plant 16, 632-642. |
| [11] | Ge LH, Jia MX, Shan HY, Gao WF, Jiang L, Cui HG, Cheng XF, Uzest M, Zhou XP, Wang AM, Li FF (2025). Viral RNA polymerase as a SUMOylation decoy inhibits RNA quality control to promote potyvirus infection. Nat Commun 16, 157. |
| [12] | Ghimire S, Tang X, Liu WG, Fu X, Zhang HH, Zhang N, Si HJ (2021). SUMO conjugating enzyme: a vital player of SUMO pathway in plants. Physiol Mol Biol Plants 27, 2421-2431. |
| [13] | Ghimire S, Tang X, Zhang N, Liu WG, Qi XH, Fu X, Si HJ (2020). Genomic analysis of the SUMO-conjugating enzyme and genes under abiotic stress in potato (Solanum tuberosum L.). Int J Genomics 2020, 9703638. |
| [14] | Ghosh S, Mellado Sanchez M, Sue-Ob K, Roy D, Jones A, Blazquez MA, Sadanandom A (2024). Charting the evolutionary path of the SUMO modification system in plants reveals molecular hardwiring of development to stress adaptation. Plant Cell 36, 3131-3144. |
| [15] | Gou MY, Huang QS, Qian WQ, Zhang ZM, Jia ZH, Hua J (2017). SUMOylation E3 ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in Arabidopsis. Mol Plant Microbe Interact 30, 334-342. |
| [16] | Hammoudi V, Fokkens L, Beerens B, Vlachakis G, Chatterjee S, Arroyo-Mateos M, Wackers PFK, Jonker MJ, van den Burg HA (2018). The Arabidopsis SUMO E3 ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth. PLoS Genet 14, e1007157. |
| [17] | Han DL, Chen C, Xia SM, Liu J, Shu J, Nguyen V, Lai JB, Cui YH, Yang CW (2021a). Chromatin-associated SUMOylation controls the transcriptional switch between plant development and heat stress responses. Plant Commun 2, 100091. |
| [18] | Han DL, Lai JB, Yang CW (2018). Research advances in functions of SUMO E3 ligases in plant growth and development. Chin Bull Bot 53, 175-184. (in Chinese) |
|
韩丹璐, 赖建彬, 阳成伟 (2018). SUMO E3连接酶在植物生长发育中的功能研究进展. 植物学报 53, 175-184.
DOI |
|
| [19] | Han DL, Lai JB, Yang CW (2021b). SUMOylation: a critical transcription modulator in plant cells. Plant Sci 310, 110987. |
| [20] | Han DL, Yu ZB, Lai JB, Yang CW (2022). Post-translational modification: a strategic response to high temperature in plants. aBIOTECH 3, 49-64. |
| [21] | Harris W, Kim S, Vӧlz R, Lee YH (2023). Nuclear effectors of plant pathogens: distinct strategies to be one step ahead. Mol Plant Pathol 24, 637-650. |
| [22] | Huang JW, Feng QY, Zheng KY, Huang JJ, Wang LB, Lai RQ, Lai JB, Yang CW (2022). An effective in vitro SUMOylation detection system for plant proteins. Chin Bull Bot 57, 490-499. (in Chinese) |
|
黄俊文, 冯琦伊, 郑凯勇, 黄俊杰, 王林博, 赖瑞强, 赖建彬, 阳成伟 (2022). 植物蛋白质SUMO化修饰体外高效检测系统. 植物学报 57, 490-499.
DOI |
|
| [23] |
Huang JW, Huang JJ, Wu JY, Zhou M, Luo SY, Jiang JM, Chen TS, Shao L, Lai JB, Yang CW (2025). A synthetic biology approach for identifying de-SUMOylation enzymes of substrates. J Integr Plant Biol 67, 1211-1213.
DOI |
| [24] | Ingole KD, Dahale SK, Bhattacharjee S (2021). Proteomic analysis of SUMO1- SUMOylome changes during defense elicitation in Arabidopsis. J Proteomics 232, 104054. |
| [25] | Jiang J, Kuo YW, Salem N, Erickson A, Falk BW (2021). Carrot mottle virus ORF4 movement protein targets plasmodesmata by interacting with the host cell SUMOylation system. New Phytol 231, 382-398. |
| [26] | Kim JG, Stork W, Mudgett MB (2013). Xanthomonas type III effector XopD deSUMOylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13, 143-154. |
| [27] | Kumar S, Zavaliev R, Wu QL, Zhou Y, Cheng J, Dillard L, Powers J, Withers J, Zhao JS, Guan ZQ, Borgnia MJ, Bartesaghi A, Dong XN, Zhou P (2022). Structural basis of NPR1 in activating plant immunity. Nature 605, 561-566. |
| [28] | Lai RQ, Jiang JM, Wang J, Du JJ, Lai JB, Yang CW (2022). Functional characterization of three maize SIZ/PIAS-type SUMO E3 ligases. J Plant Physiol 268, 153588. |
| [29] | Lai RQ, Li WL, Xu ZW, Liu W, Zeng QR, Lin WX, Jiang JM, Lai JB, Yang CW (2023). A robust method for identification of plant SUMOylation substrates in a library-based reconstitution system. Plant Commun 4, 100573. |
| [30] | Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong QQ, Ma SS, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49, 79-90. |
| [31] | Li LL, Chen JP, Sun ZT (2024a). Exploring the shared pathogenic strategies of independently evolved effectors across distinct plant viruses. Trends Microbiol 32, 1021-1033. |
| [32] | Li WL, Liu W, Xu ZW, Zhu CL, Han DL, Liao JW, Li K, Tang XY, Xie Q, Yang CW, Lai JB (2024b). Heat-induced SUMOylation differentially affects bacterial effectors in plant cells. Plant Cell 36, 2103-2116. |
| [33] | Liao XY, Sun J, Li QQ, Ding WY, Zhao BB, Wang BB, Zhou SQ, Wang HY (2023). ZmSIZ1a and ZmSIZ1b play an indispensable role in resistance against Fusarium ear rot in maize. Mol Plant Pathol 24, 711-724. |
| [34] | Lim YJ, Yoon YJ, Lee H, Choi G, Kim S, Ko J, Kim JH, Kim KT, Lee YH (2024). Nuclear localization sequence of MoHTR1, a Magnaporthe oryzae effector, for transcriptional reprogramming of immunity genes in rice. Nat Commun 15, 9764. |
| [35] |
Liu CY, Li ZG, Xing JJ, Yang J, Wang Z, Zhang H, Chen D, Peng YL, Chen XL (2018). Global analysis of SUMOylation function reveals novel insights into development and appressorium-mediated infection of the rice blast fungus. New Phytol 219, 1031-1047.
DOI PMID |
| [36] | Liu JH, Wu XY, Fang Y, Liu Y, Bello EO, Li Y, Xiong RY, Li YZ, Fu ZQ, Wang AM, Cheng XF (2023). A plant RNA virus inhibits NPR1 SUMOylation and subverts NPR1- mediated plant immunity. Nat Commun 14, 3580. |
| [37] |
Morrell R, Sadanandom A (2019). Dealing with stress: a review of plant SUMO proteases. Front Plant Sci 10, 1122.
DOI PMID |
| [38] |
Niu D, Lin XL, Kong XX, Qu GP, Cai B, Lee J, Jin JB (2019). SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12, 215-228.
DOI PMID |
| [39] | Orosa B, Yates G, Verma V, Srivastava AK, Srivastava M, Campanaro A, De Vega D, Fernandes A, Zhang CJ, Lee J, Bennett MJ, Sadanandom A (2018). SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signaling in plant innate immunity. Nat Commun 9, 5185. |
| [40] |
Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ (2011). SUMO and SUMOylation in plants. Mol Cells 32, 305-316.
DOI PMID |
| [41] | Roden J, Eardley L, Hotson A, Cao YJ, Mudgett MB (2004). Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microbe Interact 17, 633-643. |
| [42] | Rytz TC, Miller MJ, McLoughlin F, Augustine RC, Marshall RS, Juan YT, Charng YY, Scalf M, Smith LM, Vierstra RD (2018). SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30, 1077-1099. |
| [43] |
Saleh A, Withers J, Mohan R, Marqués J, Gu YN, Yan SP, Zavaliev R, Nomoto M, Tada Y, Dong XN (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169-182.
DOI PMID |
| [44] |
Sánchez-Durán MA, Dallas MB, Ascencio-Ibañez JT, Reyes MI, Arroyo-Mateos M, Ruiz-Albert J, Hanley- Bowdoin L, Bejarano ER (2011). Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection. J Virol 85, 9789-9800.
DOI PMID |
| [45] | Sang T, Xu YP, Qin GC, Zhao SS, Hsu CC, Wang PC (2024). Highly sensitive site-specific SUMOylation proteomics in Arabidopsis. Nat Plants 10, 1330-1342. |
| [46] |
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145, 119-134.
DOI PMID |
| [47] | Shao WY, Sun KW, Ma TL, Jiang HX, Hahn M, Ma ZH, Jiao C, Yin YN (2023). SUMOylation regulates low-temperature survival and oxidative DNA damage tolerance in Botrytis cinerea. New Phytol 238, 817-834. |
| [48] | Sharma M, Fuertes D, Perez-Gil J, Lois LM (2021). SUMOylation in phytopathogen interactions: balancing invasion and resistance. Front Cell Dev Biol 9, 703795. |
| [49] | Shen WZ, Liu JE, Li JF (2019). Type-II metacaspases mediate the processing of plant elicitor peptides in Arabidopsis. Mol Plant 12, 1524-1533. |
| [50] |
Skelly MJ, Malik SI, Le Bihan T, Bo Y, Jiang JH, Spoel SH, Loake GJ (2019). A role for S-nitrosylation of the SUMO-conjugating enzyme SCE1 in plant immunity. Proc Natl Acad Sci USA 116, 17090-17095.
DOI PMID |
| [51] | Soni KK, Gurjar K, Ranjan A, Sinha S, Srivastava M, Verma V (2024). Post-translational modifications control the signal at the crossroads of plant-pathogen interactions. J Exp Bot 75, 6957-6979. |
| [52] | Srivastava AK, Orosa B, Singh P, Cummins I, Walsh C, Zhang CJ, Grant M, Roberts MR, Anand GS, Fitches E, Sadanandom A (2018). SUMO suppresses the activity of the jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30, 2099-2115. |
| [53] | Srivastava M, Sadanandom A, Srivastava AK (2021). Towards understanding the multifaceted role of SUMOylation in plant growth and development. Physiol Plant 171, 77-85. |
| [54] | Tomanov K, Hardtke C, Budhiraja R, Hermkes R, Coupland G, Bachmair A (2013). Small ubiquitin-like modifier conjugating enzyme with active site mutation acts as dominant negative inhibitor of SUMO conjugation in Arabidopsis. J Integr Plant Biol 55, 75-82. |
| [55] |
Verma V, Croley F, Sadanandom A (2018). Fifty shades of SUMO: its role in immunity and at the fulcrum of the growth-defence balance. Mol Plant Pathol 19, 1537-1544.
DOI PMID |
| [56] | Verma V, Srivastava AK, Gough C, Campanaro A, Srivastava M, Morrell R, Joyce J, Bailey M, Zhang CJ, Krysan PJ, Sadanandom A (2021). SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc Natl Acad Sci USA 118, e2021351118. |
| [57] | Wu XM, Zhang BS, Zhao YL, Wu HW, Gao F, Zhang J, Zhao JH, Guo HS (2023). DeSUMOylation of a Verticillium dahliae enolase facilitates virulence by derepressing the expression of the effector VdSCP8. Nat Commun 14, 4844. |
| [58] | Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1, 639-647. |
| [59] | Xia SM, Han DL, Mo Q, Lai JB, Yang CW (2025). SUMOylation of BAK1 regulates its co-receptor function for specifically activating brassinosteroid response. Plant Commun 6, 101384. |
| [60] | Xie B, Luo MY, Li QY, Shao J, Chen DS, Somers DE, Tang DZ, Shi H (2024). NUA positively regulates plant immunity by coordination with ESD4 to deSUMOylate TPR1 in Arabidopsis. New Phytol 241, 363-377. |
| [61] | Xiong RY, Wang AM (2013). SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection. J Virol 87, 4704-4715. |
| [62] | Yu HX, Cao YJ, Yang YB, Shan JX, Ye WW, Dong NQ, Kan Y, Zhao HY, Lu ZQ, Guo SQ, Lei JJ, Liao B, Lin HX (2024). A TT1-SCE1 module integrates ubiquitination and SUMOylation to regulate heat tolerance in rice. Mol Plant 17, 1899-1918. |
| [63] |
Zavaliev R, Mohan R, Chen TY, Dong XN (2020). Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093-1108.
DOI PMID |
| [64] | Zhang C, Wu YY, Liu JE, Song B, Yu ZB, Li JF, Yang CW, Lai JB (2025). SUMOylation controls peptide processing to generate damage-associated molecular patterns in Arabidopsis. Dev Cell 60, 696-705. |
| [1] | Wu Aian, Tao Yifei, Fang Siqi, Xu Xinyue, Zhu Shanshan, Chen Shiying, Wang Tingchao, Guo Wei. Research Progress on Pathogenesis of Xanthomonas oryzae pv. oryzicola and Rice Resistance Mechanisms [J]. Chinese Bulletin of Botany, 2025, 60(5): 759-772. |
| [2] | Lumei He, Bojun Ma, Xifeng Chen. Advances on the Executor Resistance Genes in Plants [J]. Chinese Bulletin of Botany, 2024, 59(4): 671-680. |
| [3] | Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei. An Effective in Vitro SUMOylation Detection System for Plant Proteins [J]. Chinese Bulletin of Botany, 2022, 57(4): 490-499. |
| [4] | Tianxingzi Wang, Zheng Zhu, Yue Chen, Yuqing Liu, Gaowei Yan, Shan Xu, Tong Zhang, Jinjiao Ma, Shijuan Dou, Liyun Li, Guozhen Liu. Rice OsWRKY42 is a Novel Element in Xa21-mediated Resistance Pathway Against Bacterial Leaf Blight [J]. Chinese Bulletin of Botany, 2021, 56(6): 687-698. |
| [5] | Xiaomin Cui, Dongchao Ji, Tong Chen, Shiping Tian. Advances in the Studies on Molecular Mechanism of Receptor-like Protein Kinase FER Regulating Host Plant-pathogen Interaction [J]. Chinese Bulletin of Botany, 2021, 56(3): 339-346. |
| [6] | Yaning Cui, Hongping Qian, Yanxia Zhao, Xiaojuan Li. Intracellular Trafficking in Pattern Recognition Receptor-triggered Plant Immunity [J]. Chinese Bulletin of Botany, 2020, 55(3): 329-339. |
| [7] | Gaoping Qu,Jingbo Jin. Detection of SUMOylation in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 83-89. |
| [8] | Tong Zhang,Yalu Guo,Yue Chen,Jinjiao Ma,Jinping Lan,Gaowei Yan,Yuqing Liu,Shan Xu,Liyun Li,Guozhen Liu,Shijuan Dou. Expression Characterization of Rice OsPR10A and Its Function in Response to Drought Stress [J]. Chinese Bulletin of Botany, 2019, 54(6): 711-722. |
| [9] | Tingting Shan,Xiaomei Chen,Shunxing Guo,Lixia Tian,Lin Yan,Xin Wang. Advances in Molecular Regulation of Sphingolipids in Plant-fungus Interactions [J]. Chinese Bulletin of Botany, 2019, 54(3): 396-404. |
| [10] | Jing Zhang,Suiwen Hou. Role of Post-translational Modification of Proteins in ABA Signaling Transduction [J]. Chinese Bulletin of Botany, 2019, 54(3): 300-315. |
| [11] | Yaqiong Liu,Suiwen Hou. Research Progress in Protein Phosphorylation in Plant-pathogen Interactions [J]. Chinese Bulletin of Botany, 2019, 54(2): 168-184. |
| [12] | Han Danlu, Lai Jianbin, Yang Chengwei. Research Advances in Functions of SUMO E3 Ligases in Plant Growth and Development [J]. Chinese Bulletin of Botany, 2018, 53(2): 175-184. |
| [13] | Ronggai Li, Yanmei Lu, Yueying Wang, Baoqiang Wang, Wei Song, Wenying Zhang. Molecular Study on Maize Rough Dwarf Disease: A Review [J]. Chinese Bulletin of Botany, 2017, 52(3): 375-387. |
| [14] | Dongchao Ji, Kai Song, Jingjing Xing, Tong Chen, Shiping Tian. Studies of Innate Immunity Mediated by Lysin Motif Protein and Its Signaling Priming [J]. Chinese Bulletin of Botany, 2015, 50(5): 628-636. |
| [15] | Xinli Zan, Ying Gao, Yuling Chen, Kaijun Zhao. Pathogen-responsive Cis-acting Elements and Their Interactive Transcription Factors [J]. Chinese Bulletin of Botany, 2013, 48(2): 219-229. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||