

Chinese Bulletin of Botany ›› 2025, Vol. 60 ›› Issue (1): 49-61.DOI: 10.11983/CBB24019 cstr: 32102.14.CBB24019
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Liuqing Yang,†, Jin Wang,†, Jingli Yan, Qinqin Chen, Haokun Cheng, Chun Li, Peiyu Zhao, Bo Yang, Yuanqing Jiang*( )
)
Received:2024-02-05
Accepted:2024-08-20
Online:2025-01-10
Published:2024-08-22
Contact:
* E-mail: About author:†These authors contributed equally to this paper
Liuqing Yang, Jin Wang, Jingli Yan, Qinqin Chen, Haokun Cheng, Chun Li, Peiyu Zhao, Bo Yang, Yuanqing Jiang. Analysis of Expression Characteristics and Identification of Interaction Proteins of BnaABF2 Transcription Factor in Brassica napus[J]. Chinese Bulletin of Botany, 2025, 60(1): 49-61.
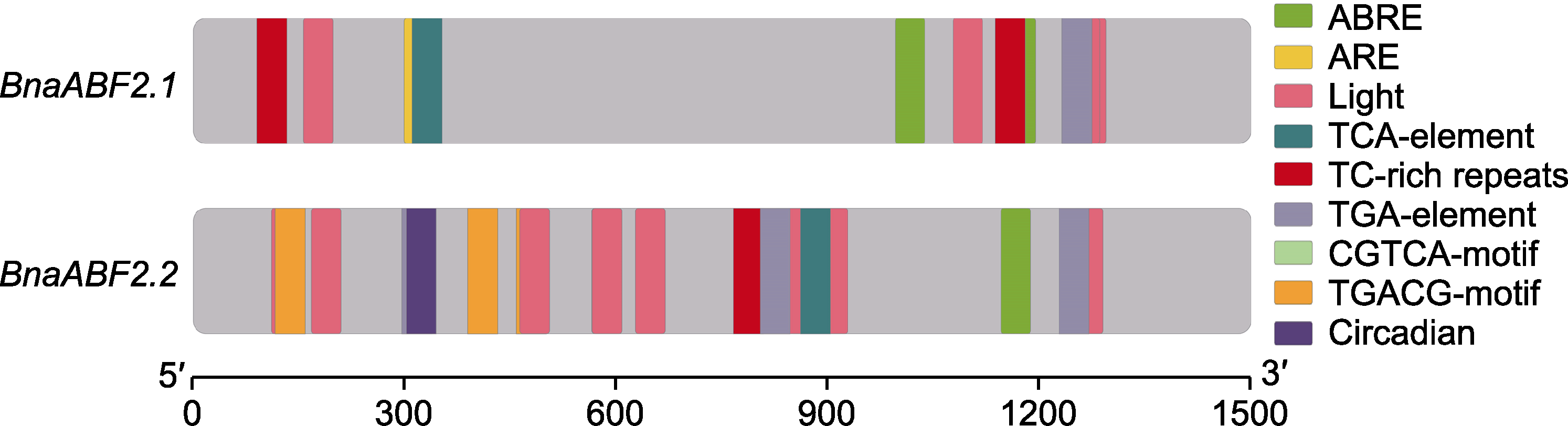
Figure 1 Cis-elements of two BnaABF2 promoters ABRE: Abscisic acid responsiveness element; ARE: Anaerobic induction element; Light: Light responsiveness element; TCA- element: Salicylic acid responsiveness element; TC-rich repeats: Defense and stress responsiveness element; TGA-element: Auxin responsiveness element; CGTCA-motif: MeJA-responsiveness motif; TGACG-motif: Jasmonic acid responsiveness motif; Circadian: Circadian rhythm
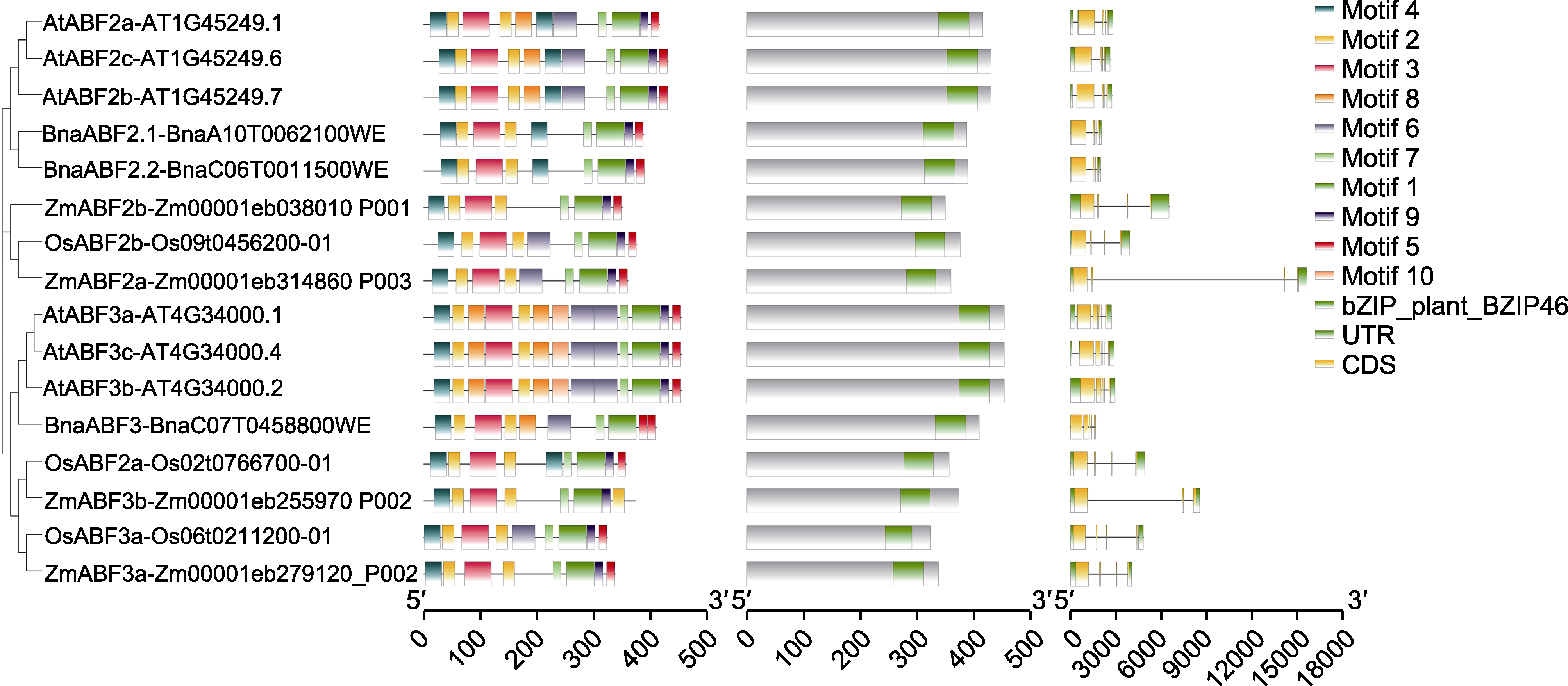
Figure 2 Sequence alignment of conserved domains of BnaABF2 with ABF from other species Left: Protein conserved motifs of ABF2/3 alignment; Middle: ABF2/3 protein conserved domain alignment; Right: Transcript structure alignment of coding region (CDS) and untranslated region (UTR) of ABF2/3.
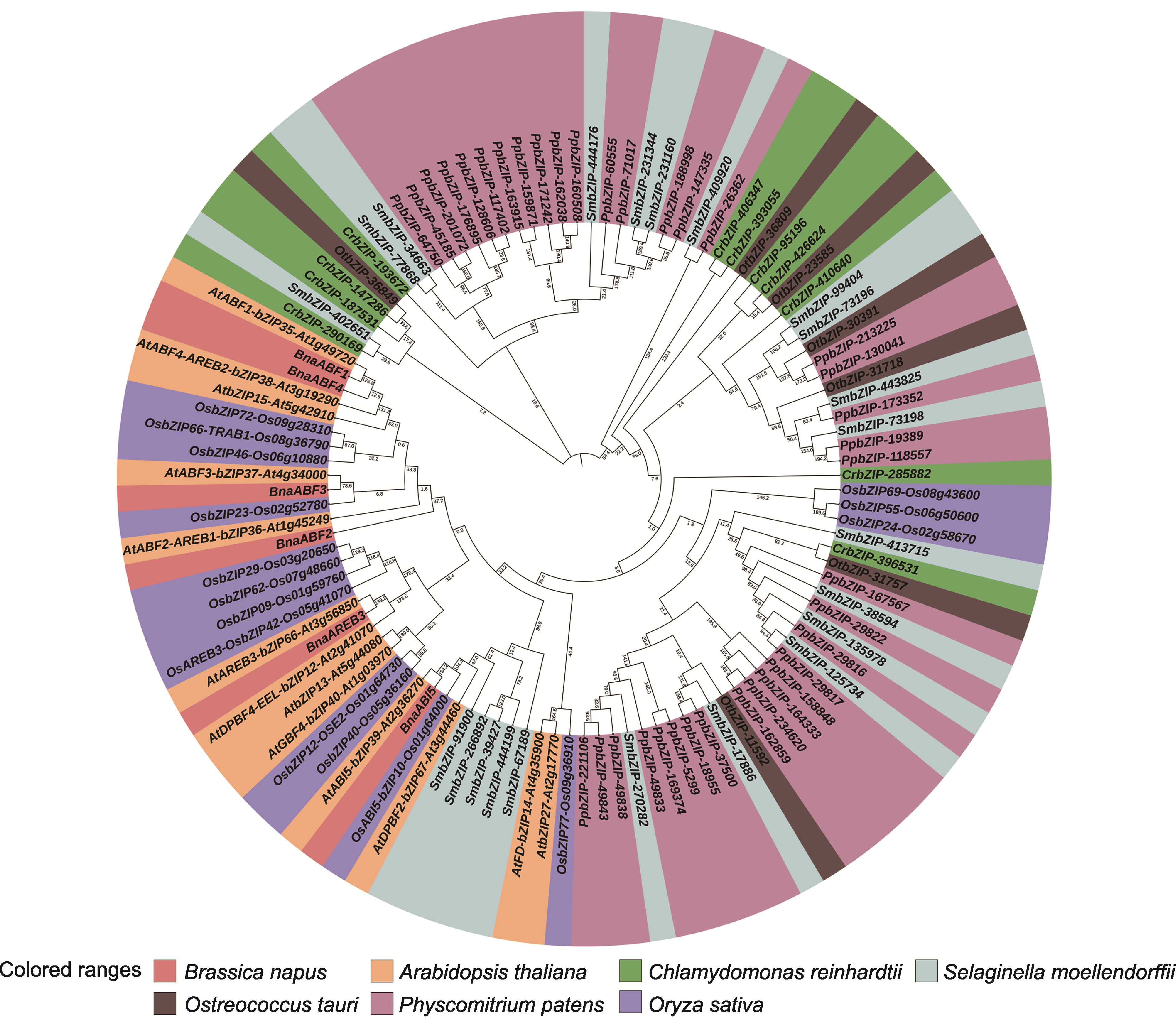
Figure 3 A phylogenetic tree of BnaABF2 and ABF proteins from other species The branches of the evolutionary tree showed genetic distance with a confidence level of 100.
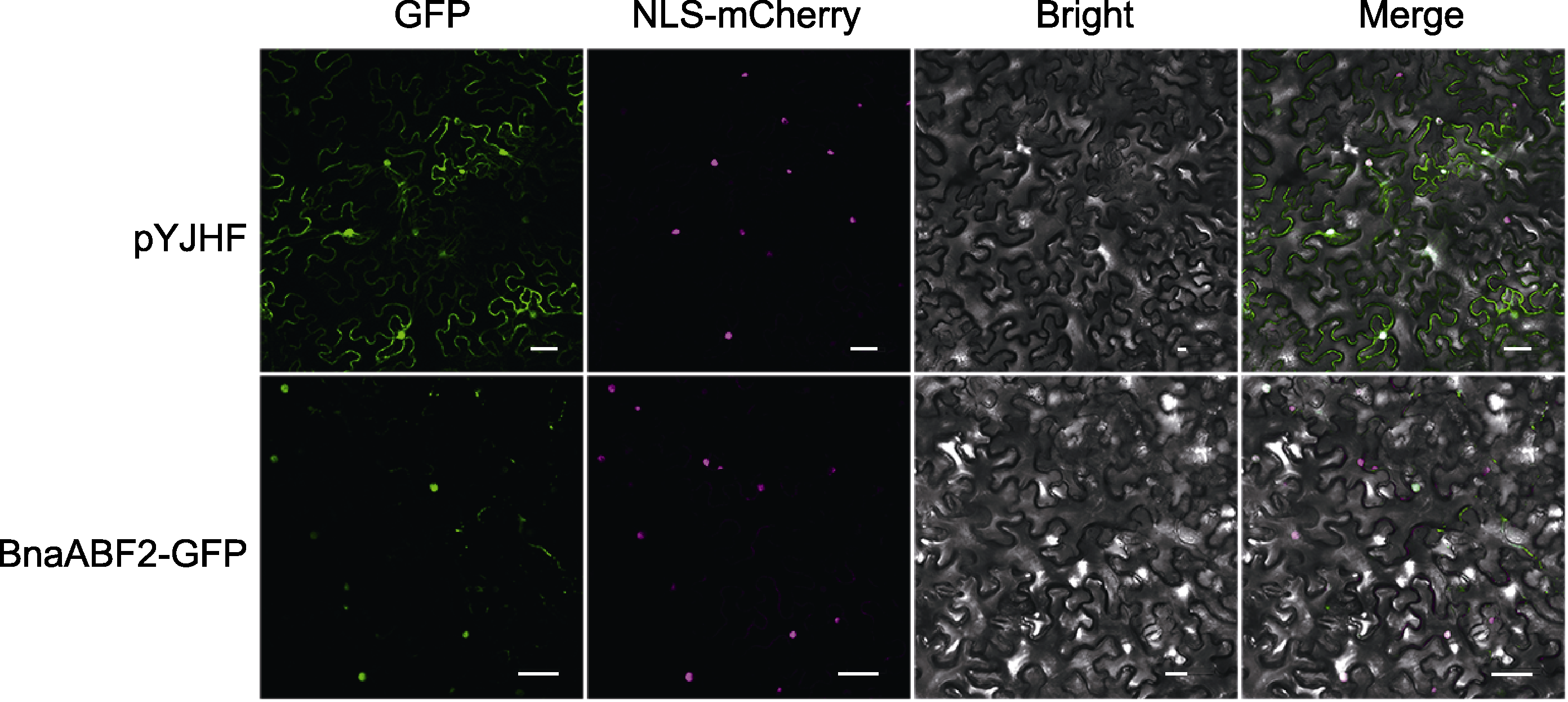
Figure 4 Subcellular localization of BnaABF2 pYJHF-BnaABF2 or pYJHF (control) plasmid were infiltrated into tobacco leaves, and fluorescence signals were observed two days later. GFP: Green fluorescent protein signal; NLS-mCherry: Nuclear localization marker; Bright: Bright field; Merge: Superimposed field. Bars=50 μm
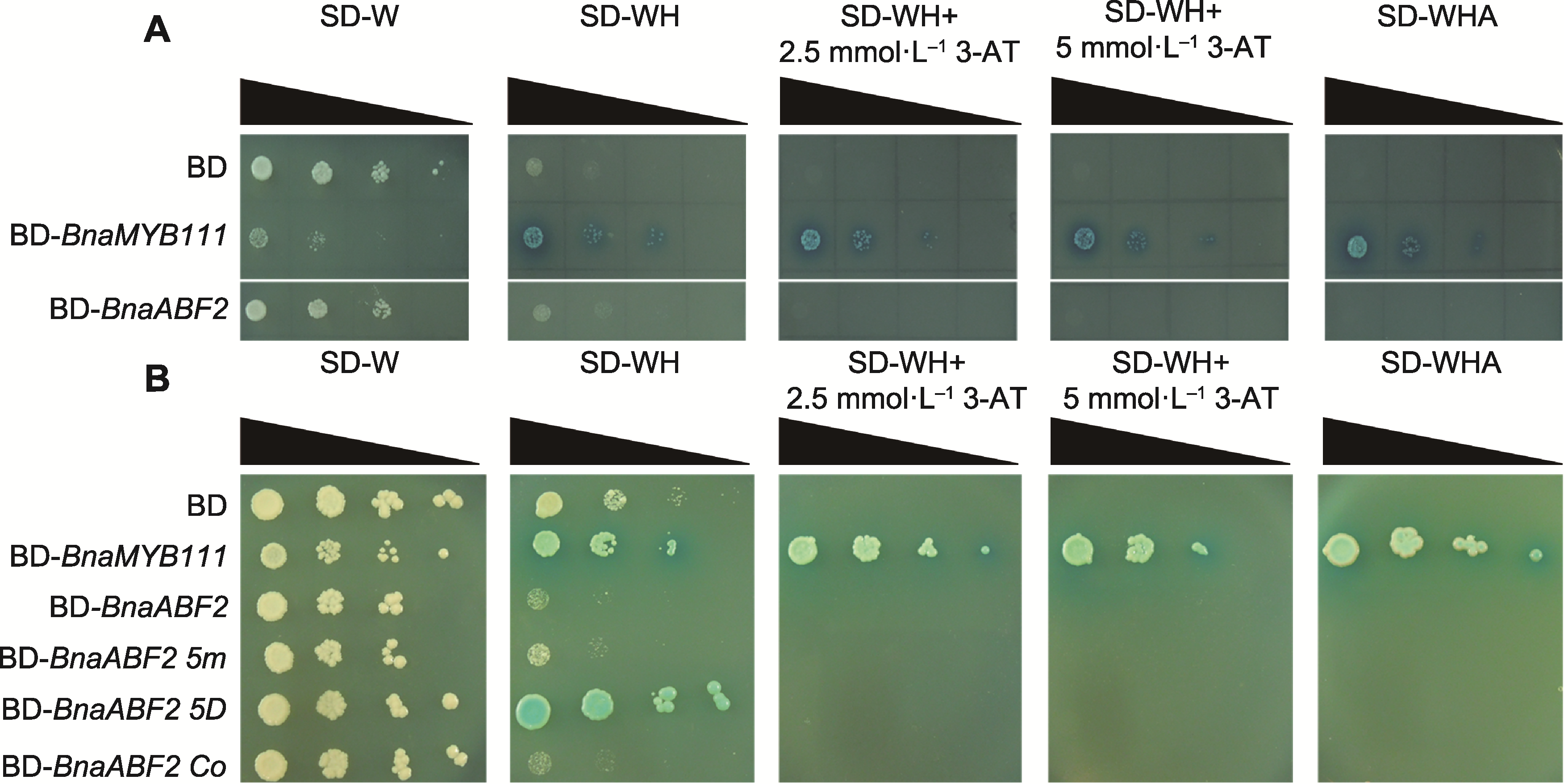
Figure 5 Analysis of transcriptional activity of BnaABF2 in yeast system BD (negative control), BD-BnaABF2, BD-BnaMYB111 (positive control), BD-BnaABF2 5m, BD-BnaABF2 5D and BD-BnaABF2 Co were transferred into yeast for titration experiment. The dilution gradients of yeast cells were marked with black triangles.
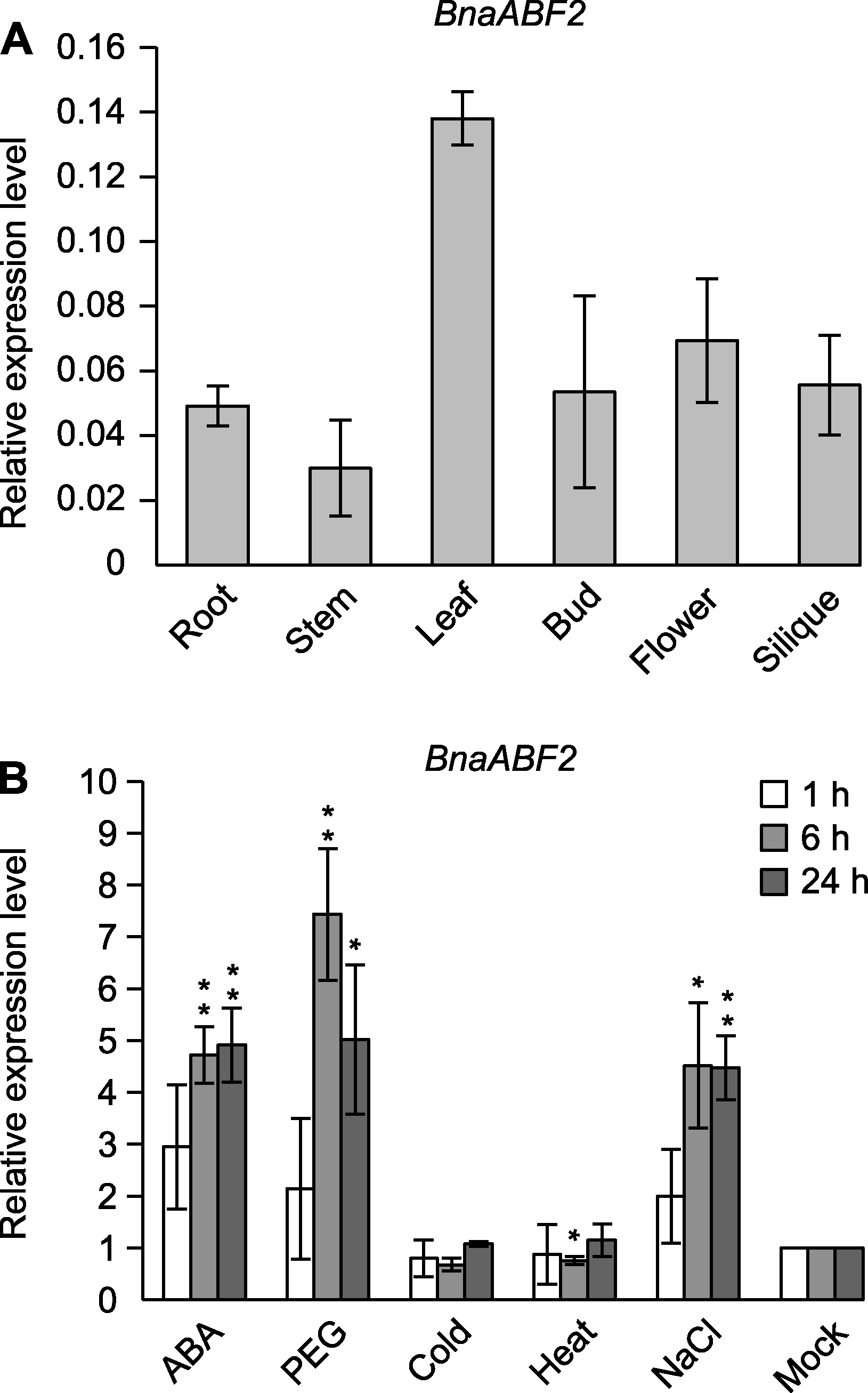
Figure 6 Expression patterns of BnaABF2 gene in different tissues and under different stresses and plant growth regulator treatments (A) The expression of BnaABF2 in different tissues; (B) The expression of BnaABF2 under various stresses and plant growth regulator treatments, the treatments included 50 μmol·L-1 ABA, 15% PEG8000, cold (4°C), heat (38°C) and salinity (200 mmol·L-1 NaCl). Values are means±SE. The significant differences in t-test were indicated by asterisk, * P<0.05; ** P<0.01
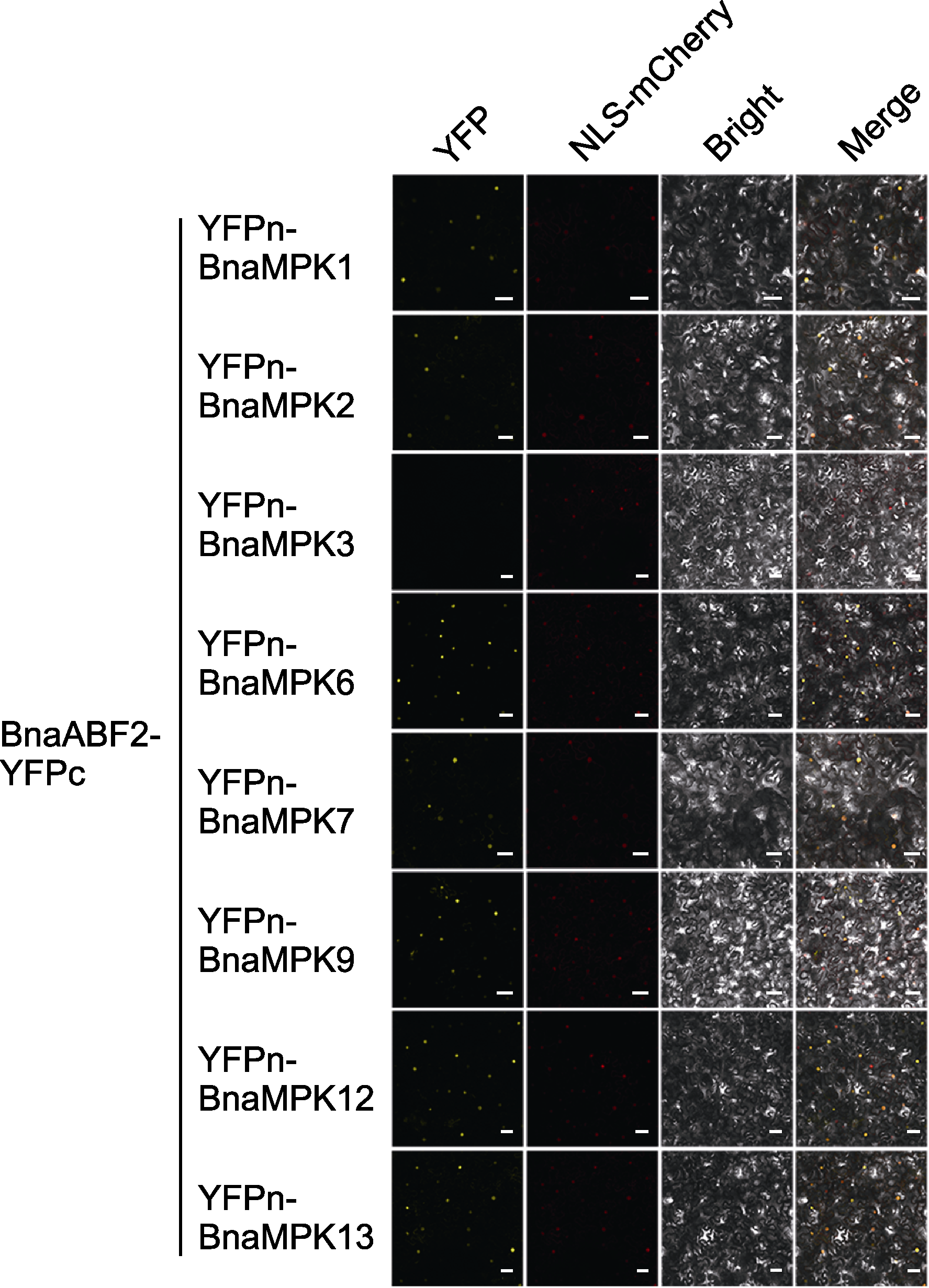
Figure 7 Screening of interaction between BnaABF2 and BnaMPK proteins through bimolecular fluorescence complementation assay The plasmids of BnaABF2-YFPc and YFPn-BnaMPK1/2/6/ 7/9/12/13 and NLS-mCherry were co-transfected into tobacco (28 d). Four days later, the fluorescence signal was observed. YFPn: Yellow fluorescent protein N-terminal sequence; YFPc: Yellow fluorescent protein C-terminal sequence; YFP: Yellow fluorescent protein signal; NLS-mCherry: Nuclear localization red fluorescence signal; Bright: Bright field; Merge: Superimposed field. Bars=50 μm
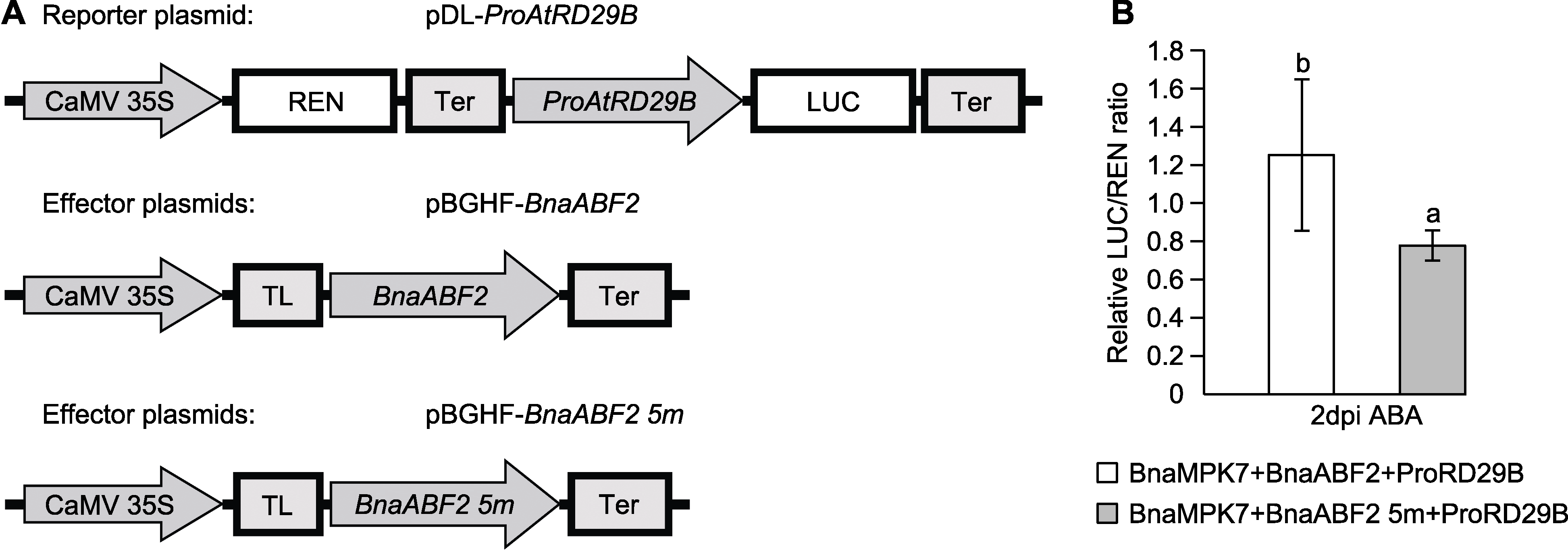
Figure 8 Dual-luciferase reporter system detects the effect of BnaABF2 on the transcriptional activity of AtRD29B promoter under abscisic acid (ABA) treatment (A) A schematic diagram of the reporter and effector; (B) The LUC/REN ratio is used to indicate the ability of the effector to activate the reporter (dpi: Days post-infiltration). Different lowercase letters indicate significant differences between different treatments (P<0.05).
| [1] | Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645-2663. |
| [2] | Agarwal PK, Jha B (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol Plant 54, 201-212. |
| [3] |
Chalhoub B, Denoeud F, Liu SY, Parkin IA, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950-953.
DOI PMID |
| [4] |
Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000). ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275, 1723-1730.
DOI PMID |
| [5] | Colcombet J, Hirt H (2008). Arabidopsis MAPKs: a complex signaling network involved in multiple biological processes. Biochem J 413, 217-226. |
| [6] | Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, dit Frey NF, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, Marcote MJ, Hirt H, Colcombet J (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82, 232-244. |
| [7] | Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470-3488. |
| [8] | Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103, 1988-1993. |
| [9] |
Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK (2002). The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130, 837-846.
DOI PMID |
| [10] | Kagale S, Links MG, Rozwadowski K (2010). Genome- wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152, 1109-1134. |
| [11] |
Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40, 75-87.
DOI PMID |
| [12] | Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017). Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45, 6613-6627. |
| [13] |
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44, 939-949.
DOI PMID |
| [14] | Li QY, Liu C, He L, Peng S, Ma JY, Hu ZY, Liu HB (2025). Cloning and functional analysis of BnaA02.CPSF6 gene from Brassica napus. Chin Bull Bot 60, 62-73. (in Chinese) |
|
李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波 (2025). 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析. 植物学报 60, 62-73.
DOI |
|
| [15] | Li SN, Wang WY, Gao JL, Yin KQ, Wang R, Wang CC, Petersen M, Mundy J, Qiu JL (2016). MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28, 2866-2883. |
| [16] | Li YH, Liu K, Tong GL, Xi C, Liu J, Zhao HP, Wang YD, Ren DT, Han SC (2022). MPK3/MPK6-mediated phosphorylation of ERF72 positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin biosynthesis enzymes. J Exp Bot 73, 413-428. |
| [17] | Liang WW, Yang B, Yu BJ, Zhou ZL, Li C, Jia M, Sun Y, Zhang Y, Wu FF, Zhang HF, Wang BY, Deyholos MK, Jiang YQ (2013). Identification and analysis of MKK and MPK gene families in canola (Brassica napus L.). BMC Genomics 14, 392. |
| [18] | Mao GH, Meng XZ, Liu YD, Zheng ZY, Chen ZX, Zhang SQ (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23 1639-1653. |
| [19] | Niu FF, Wang C, Yan JL, Guo XH, Wu FF, Yang B, Deyholos MK, Jiang YQ (2016). Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol Biol 92, 89-104. |
| [20] |
Rodriguez MSC, Petersen M, Mundy J (2010). Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61, 621-649.
DOI PMID |
| [21] | Sun Y, Wang C, Yang B, Wu FF, Hao XY, Liang WW, Niu FF, Yan JL, Zhang HF, Wang BY, Deyholos MK, Jiang YQ (2014). Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J Exp Bot 65, 2171-2188. |
| [22] |
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid- dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97, 11632-11637.
DOI PMID |
| [23] | Wang HC, Ngwenyama N, Liu YD, Walker JC, Zhang SQ (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63-73. |
| [24] |
Xu J, Zhang SQ (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20, 56-64.
DOI PMID |
| [25] | Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61, 672-685. |
| [26] | Zhang HF (2019). Exploration of the Mechanisms of Two Calcium-Associated Protein Kinases Regulating ABA Signaling Transduction in Arabidopsis. PhD dissertation. Yang-ling: Northwest A&F University. (in Chinese) |
| 张翰风 (2019). 拟南芥中两个钙相关蛋白激酶调控ABA信号转导的机制研究. 博士论文. 杨凌: 西北农林科技大学. | |
| [27] |
Zhang MM, Zhang SQ (2022). Mitogen-activated protein kinase cascades in plant signaling. J Integr Plant Biol 64, 301-341.
DOI |
| [28] | Zhao BY, Hu YF, Li JJ, Yao X, Liu KD (2016). BnaABF2, a bZIP transcription factor from rapeseed (Brassica napus L.), enhances drought and salt tolerance in transgenic Arabidopsis. Bot Stud 57, 12. |
| [29] | Zhou YP, Yan JH, Tian CE (2022). Research progress on the regulatory mechanisms of ABA signal transduction in guard cells. Chin Bull Bot 57, 684-696. (in Chinese) |
|
周玉萍, 颜嘉豪, 田长恩 (2022). 保卫细胞中ABA信号调控机制研究进展. 植物学报 57, 684-696.
DOI |
|
| [30] | Zhu JK (2016). Abiotic stress signaling and responses in plants. Cell 167, 313-324. |
| [1] | Qingyang Li, Cui Liu, Li He, Shan Peng, Jiayin Ma, Ziyi Hu, Hongbo Liu. Cloning and Functional Analysis of the BnaA02.CPSF6 Gene from Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 62-73. |
| [2] | Yi Song, Hanghang Chen, Xin Cui, Zhifeng Lu, Shipeng Liao, Yangyang Zhang, Xiaokun Li, Rihuan Cong, Tao Ren, Jianwei Lu. Potassium Nutrient Status-mediated Leaf Growth of Oilseed Rape (Brassica napus) and Its Effect on Phyllosphere Microorganism [J]. Chinese Bulletin of Botany, 2024, 59(1): 54-65. |
| [3] | Wang Wenguang, Wang Yonghong. Century-old Hypothesis Finally Revealed: the Shuttling LAZY Proteins “Awaken” Gravity Sensing in Planta [J]. Chinese Bulletin of Botany, 2023, 58(5): 677-681. |
| [4] | Zhang Yingchuan, Wu Xiaomingyu, Tao Baolong, Chen Li, Lu Haiqin, Zhao Lun, Wen Jing, Yi Bin, Tu Jinxing, Fu Tingdong, Shen Jinxiong. Bna-miR43 Mediates the Response of Drought Tolerance in Brassica napus [J]. Chinese Bulletin of Botany, 2023, 58(5): 701-711. |
| [5] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [6] | Min Song,Yao Zhang,Liying Wang,Xiangyong Peng. Genome-wide Identification and Phylogenetic Analysis of Zinc Finger Homeodomain Family Genes in Brassica napus [J]. Chinese Bulletin of Botany, 2019, 54(6): 699-710. |
| [7] | Jia Guo,Yansu Li,Chaoxing He,Yan Yan,Xianchang Yu. Establishing a High-efficiency Regeneration System in Pumpkin (Cucurbita moschata) [J]. Chinese Bulletin of Botany, 2019, 54(4): 539-546. |
| [8] | Liu Kaige, Qi Shuanghui, Duan Shaowei, Li Dong, Jin Changyu, Gao Chenhao, Liu Mingxun Chen Xuanxia. Functional Analysis of Brassica napus BnTTG1-1 Gene [J]. Chinese Bulletin of Botany, 2017, 52(6): 713-722. |
| [9] | Gao Huhu, Zhang Yunxiao, Hu Shengwu, Guo Yuan. Genome-wide Survey and Phylogenetic Analysis of MADS-box Gene Family in Brassica napus [J]. Chinese Bulletin of Botany, 2017, 52(6): 699-712. |
| [10] | Jia Ledong, Li Shimeng, Xu Daixiang, Qu Cunmin, Li Jiana, Wang Rui. Bioinformatics Analysis of BnMYB80 Genes in Brassica napus [J]. Chinese Bulletin of Botany, 2016, 51(5): 620-630. |
| [11] | Ziyang Min, Han Li, Tian Zou, Long Tong, Juan Cheng, Xiaowu Sun. Studies of in Vitro Culture and Plant Regeneration of Unfertilized Ovary of Pumpkin [J]. Chinese Bulletin of Botany, 2016, 51(1): 74-80. |
| [12] | Yue Liu, Yuejia Yin, Chongyang Liang, Dianshuai Huang, Yang Wang, Yanzhi Liu, Yao Dou, Shudan Feng, Dongyun Hao. Using 3D-SIM Structure Illumination Microscope to Localize Proteins in Plant Subcellular Compartments [J]. Chinese Bulletin of Botany, 2015, 50(4): 495-503. |
| [13] | Shixuan Chen, Zhennan Zhang, Bo Wang, Yan Zhu, Yuehua Gong, Dongmei Sun, Xin Deng. Cloning, Expression and Functional Analysis of a J-domain Protein- coding Gene, BhDNAJC2, from the Resurrection Plant Boea hygrometrica [J]. Chinese Bulletin of Botany, 2015, 50(2): 180-190. |
| [14] | YANG Chun, TAN Tai-Long, YU Jia-Ling, LIAO Qiong, ZHANG Xiao-Long, ZHANG Zhen-Hua, SONG Hai-Xing, GUAN Chun-Yun. Effects of atmospheric CO2 enrichment on phloem sap composition and root nitrogen accumulation in oilseed rape [J]. Chin J Plant Ecol, 2014, 38(7): 776-784. |
| [15] | Fang Wang. RabD2b Protein N121I Mutation Affects the Subcellular Localization and Functions in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2014, 49(6): 653-662. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||