

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (6): 699-710.DOI: 10.11983/CBB19055 cstr: 32102.14.CBB19055
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Min Song( ),Yao Zhang,Liying Wang,Xiangyong Peng
),Yao Zhang,Liying Wang,Xiangyong Peng
Received:2019-03-25
Accepted:2019-07-26
Online:2019-11-01
Published:2020-07-09
Contact:
Min Song
Min Song,Yao Zhang,Liying Wang,Xiangyong Peng. Genome-wide Identification and Phylogenetic Analysis of Zinc Finger Homeodomain Family Genes in Brassica napus[J]. Chinese Bulletin of Botany, 2019, 54(6): 699-710.
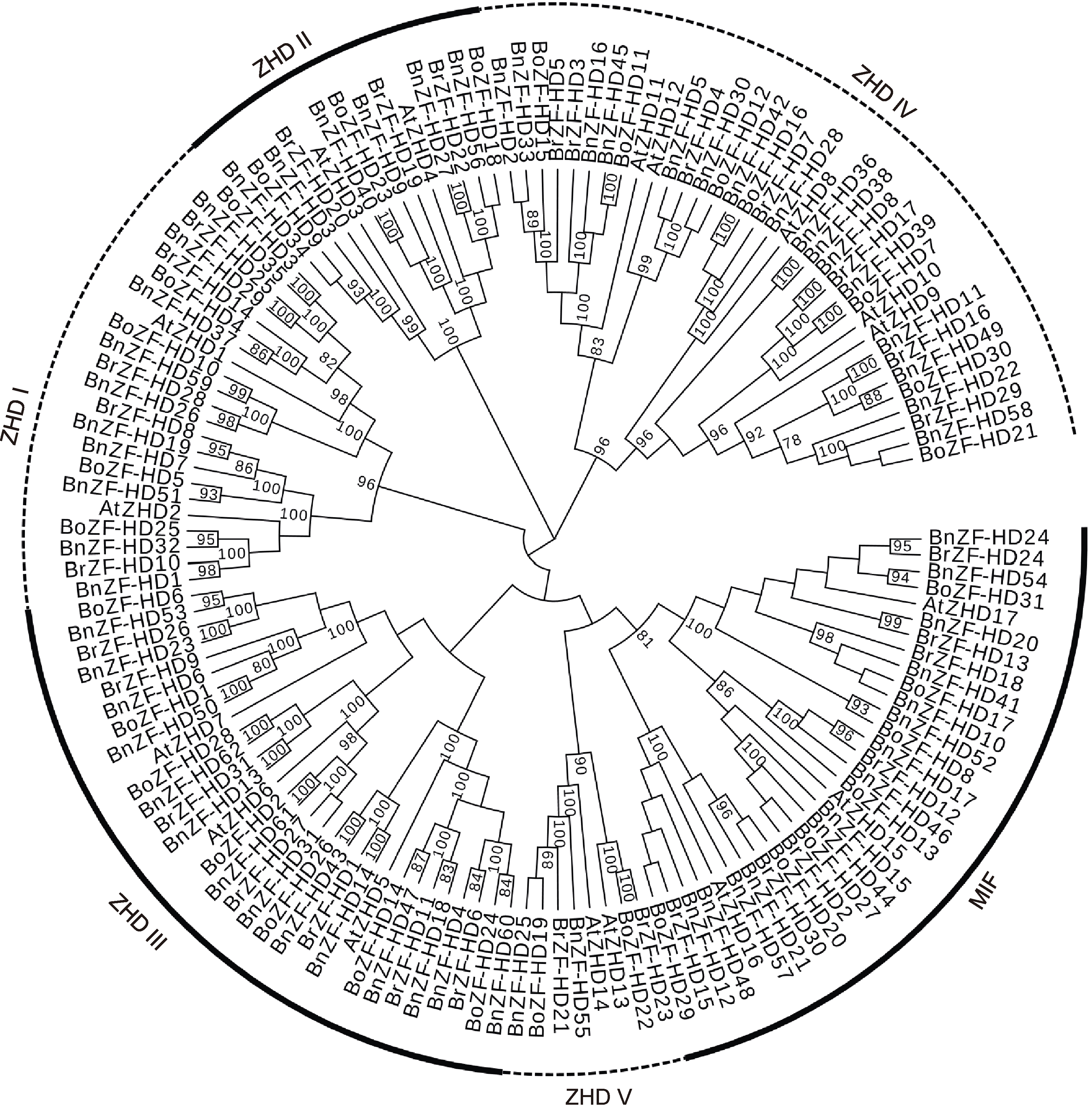
Figure 1 Phylogenetic analysis of Brassica napus, Arabidopsis thaliana, B. rapa and B. oleracea ZF-HD proteins This phylogenetic unrooted tree was constructed by the neighbor-joining (NJ) method with MEGA 7.0, with 1 000 bootstrap replicates.
| [1] | 高虎虎, 张云霄, 胡胜武, 郭媛 ( 2017). 甘蓝型油菜MADS- box基因家族的鉴定与系统进化分析. 植物学报 52, 699-712. |
| [2] | Abdullah M, Cheng X, Cao YP, Su XQ, Manzoor MA, Gao JS, Cai YP, Lin Y ( 2018). Zinc finger-homeodomain transcriptional factors (ZHDs) in upland cotton ( Gossypium hirsutum ): genome-wide identification and expression analysis in fiber development. Front Genet 9, 357. |
| [3] | Ariel FD, Manavella PA, Dezar CA, Chan RL ( 2007). The true story of the HD-Zip family. Trends Plant Sci 12, 419-426. |
| [4] | Bailey TL, Johnson J, Grant CE, Noble WS ( 2015). The MEME suite. Nucleic Acids Res 43, W39-W49. |
| [5] | Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL ( 2002). The Pfam protein families database. Nucleic Acids Res 30, 276-280. |
| [6] | Bhattacharjee A, Ghangal R, Garg R, Jain M ( 2015). Genome-wide analysis of homeobox gene family in legumes: identification, gene duplication and expression profiling. PLoS One 10, e0119198. |
| [7] | Bhattacharjee A, Jain M (2013). omeobox genes as potential candidates for crop improvement under abiotic stress. In: Tuteja N, Singh Gill S, eds. Plant Acclimation to Environmental Stress. New York: Springer. pp. 163-176. |
| [8] | Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P ( 2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950-953. |
| [9] | Chen CJ, Xia R, Chen H, He YH ( 2018). TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv [2018-03-27]. . |
| [10] | Cheng F, Liu SY, Wu J, Fang L, Sun SL, Liu B, Li PX, Hua W, Wang XW ( 2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11, 136. |
| [11] | Cheng F, Wu J, Fang L, Wang XW ( 2012). Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci 3, 198. |
| [12] | Deng X, Phillips J, Meijer AH, Salamini F, Bartels D ( 2002). Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Mol Biol 49, 601-610. |
| [13] | Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A ( 2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44, D279-D285. |
| [14] | He ZL, Zhang HK, Gao SH, Lercher MJ, Chen WH, Hu SN ( 2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44, W236-W241. |
| [15] | Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G ( 2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296-1297. |
| [16] | Hu W, DePamphilis CW, Ma H ( 2008). Phylogenetic analysis of the plant-specific Zinc finger-Homeobox and Mini zinc finger gene families. J Integr Plant Biol 50, 1031-1045. |
| [17] | Hu W, Ma H ( 2006). Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45, 399-422. |
| [18] | Jain M, Tyagi AK, Khurana JP ( 2008). Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J 275, 2845-2861. |
| [19] | Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, Gao G ( 2017a). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45, D1040-D1045. |
| [20] | Jin XL, Ren J, Nevo E, Yin XG, Sun DF, Peng JH ( 2017b). Divergent evolutionary patterns of NAC transcription factors are associated with diversification and gene duplications in angiosperm. Front Plant Sci 8, 1156. |
| [21] | Jørgensen JE, Grønlund M, Pallisgaard N, Larsen K, Marcker KA, Jensen EØ ( 1999). A new class of plant homeobox genes is expressed in specific regions of determinate symbiotic root nodules. Plant Mol Biol 40, 65-77. |
| [22] | Khadiza K, Nath UK, Robin AHK, Park JI, Lee DJ, Kim MB, Kim CK, Lim LB, Nou IS, Chung MY ( 2017). Genome-wide analysis and expression profiling of zinc finger homeodomain ( ZHD ) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genomics 18, 695. |
| [23] | Krishna SS, Majumdar I, Grishin NV ( 2003). SURVEY AND SUMMARY: structural classification of zinc fingers. Nucleic Acids Res 31, 532-550. |
| [24] | Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA ( 2009). Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639-1645. |
| [25] | Kumar S, Stecher G, Tamura K ( 2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870-1874. |
| [26] | Liu SY, Liu YM, Yang XH, Tong CB, Edwards D, Parkin IAP, Zhao MX, Ma JX, Yu JY, Huang SM, Wang XY, Wang JY, Lu K, Fang ZY, Bancroft I, Yang TJ, Hu Q, Wang XF, Yue Z, Li HJ, Yang LF, Wu J, Zhou Q, Wang WX, King GJ, Pires JC, Lu CX, Wu ZY, Sampath P, Wang Z, Guo H, Pan SK, Yang LM, Min JM, Zhang D, Jin DC, Li WS, Belcram H, Tu JX, Guan M, Qi CK, Du DZ, Li JN, Jiang LC, Batley J, Sharpe AG, Park BS, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong CH, Wang L, Li JP, Hu ZY, Zhuang M, Huang Y, Huang JY, Shi JQ, Mei DS, Liu J, Lee TH, Wang JP, Jin HZ, Li ZY, Li X, Zhang JF, Xiao L, Zhou YM, Liu ZS, Liu XQ, Qin R, Tang X, Liu WB, Wang YP, Zhang YY, Lee J, Kim HH, Denoeud F, Xu X, Liang XM, Hua W, Wang XW, Wang J, Chalhoub B, Paterson AH ( 2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5, 3930. |
| [27] | Mackay JP, Crossley M ( 1998). Zinc fingers are sticking together. Trends Biochem Sci 23, 1-4. |
| [28] | Mun JH, Kwon SJ, Yang TJ, Seol YJ, Jin M, Kim JA, Lim MH, Kim JS, Baek S, Choi BS, Yu HJ, Kim DS, Kim N, Lim KB, Lee SI, Hahn JH, Lim YP, Bancroft I, Park BS ( 2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol 10, 211. |
| [29] | Nam J, Nei M ( 2005). Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol 22, 2386-2394. |
| [30] | Nekrutenko A, Makova KD, Li WH ( 2002). The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res 12, 198-202. |
| [31] | Park HC, Kim ML, Lee SM, Bahk JD, Yun DJ, Lim CO, Hong JC, Lee SY, Cho MJ, Chung WS ( 2007). Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res 35, 3612-3623. |
| [32] | Schultz J, Milpetz F, Bork P, Ponting CP ( 1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95, 5857-5864. |
| [33] | Takatsuji H ( 1999). Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol 39, 1073-1078. |
| [34] | Tan QKG, Irish VF ( 2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol 140, 1095-1108. |
| [35] | Tran LSP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K ( 2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49, 46-63. |
| [36] | Walther D, Brunnemann R, Selbig J ( 2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet 3, e11. |
| [37] | Wang H, Yin XJ, Li XQ, Wang L, Zheng Y, Xu XZ, Zhang YC, Wang XP ( 2014). Genome-wide identification, evolution and expression analysis of the grape ( Vitis vinifera L.) zinc finger-homeodomain gene family. Int J Mol Sci 15, 5730-5748. |
| [38] | Wang W, Wu P, Li Y, Hou X ( 2016). Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol Gen Genomics 291, 1451-1464. |
| [39] | Wang XW, Wang HZ, Wang J, Sun RF, Wu J, Liu SY, Bai YQ, Mun JH, Bancroft I, Cheng F, Huang SW, Li XX, Hua W, Wang JY, Wang XY, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu BH, Li B, Liu B, Tong CB, Song C, Duran C, Peng CF, Geng CY, Koh C, Lin CY, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang HB, Wang HP, Belcram H, Zhou HL, Hirakawa H, Abe H, Guo H, Wang H, Jin HZ, Parkin IAP, Batley J, Kim JS, Just J, Li JW, Xu JH, Deng J, Kim JA, Li JP, Yu JY, Meng JL, Wang JP, Min JM, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao MX, Jin MN, Ramchiary N, Drou N, Berkman PJ, Cai QL, Huang QF, Li RQ, Tabata S, Cheng SF, Zhang S, Zhang SJ, Huang SM, Sato S, Sun SL, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li YR, Du YC, Liao YC, Lim Y, Narusaka Y, Wang YP, Wang ZY, Li ZY, Wang ZW, Xiong ZY, Zhang ZH ( 2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43, 1035-1039. |
| [40] | Windhövel A, Hein I, Dabrowa R, Stockhaus J ( 2001). Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol 45, 201-214. |
| [41] | Yamaguchi-Shinozaki K, Shinozaki K ( 2005). Organization of cis -acting regulatory elements in osmotic-and cold- stress-responsive promoters. Trends Plant Sci 10, 88-94. |
| [42] | Yang JH, Liu DY, Wang XW, Ji CM, Cheng F, Liu BN, Hu ZY, Chen S, Pental D, Ju YH, Yao P, Li XM, Xie K, Zhang JH, Wang JL, Liu F, Ma WW, Shopan J, Zheng HK, Mackenzie SA, Zhang MF ( 2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48, 1225-1232. |
| [1] | Xiaolin Chu, Quanguo Zhang. A review of experimental evidence for the evolutionary speed hypothesis [J]. Biodiv Sci, 2025, 33(4): 25019-. |
| [2] | Bailong Zhao, Yeliang Li, Yufei Wang, Bin Sun. New Leaf Classification of Mahonia (Berberidaceae) [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [3] | Linfeng Xia, Rui Li, Haizheng Wang, Daling Feng, Chunyang Wang. Research Advances and Prospects in Charophytes Genomics [J]. Chinese Bulletin of Botany, 2025, 60(2): 271-282. |
| [4] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [5] | Qingyang Li, Cui Liu, Li He, Shan Peng, Jiayin Ma, Ziyi Hu, Hongbo Liu. Cloning and Functional Analysis of the BnaA02.CPSF6 Gene from Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 62-73. |
| [6] | Liuqing Yang, Jin Wang, Jingli Yan, Qinqin Chen, Haokun Cheng, Chun Li, Peiyu Zhao, Bo Yang, Yuanqing Jiang. Analysis of Expression Characteristics and Identification of Interaction Proteins of BnaABF2 Transcription Factor in Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 49-61. |
| [7] | Nan Chen, Quan-Guo Zhang. The experimental evolution approach [J]. Biodiv Sci, 2024, 32(9): 24171-. |
| [8] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [9] | Xiang Song, Luyao Wang, Boxiao Fu, Shuangda Li, Yuanyuan Wei, Yan Hong, Silan Dai. Advances in Identification and Synthesis of Promoter Elements in Higher Plants [J]. Chinese Bulletin of Botany, 2024, 59(5): 691-708. |
| [10] | Yingli Cai, Hongge Zhu, Jiaxin Li. Biodiversity conservation in China: Policy evolution, main measures and development trends [J]. Biodiv Sci, 2024, 32(5): 23386-. |
| [11] | Rui Qu, Zhenjun Zuo, Youxin Wang, Liangjian Zhang, Zhigang Wu, Xiujuan Qiao, Zhong Wang. The biogeochemical niche based on elementome and its applications in different ecosystems [J]. Biodiv Sci, 2024, 32(4): 23378-. |
| [12] | Jixuan Yang, Xuefei Wang, Hongya Gu. Genetic Basis of Flowering Time Variations in Tibetan Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2024, 59(3): 373-382. |
| [13] | Zhi Yang, Yong Yang. Research Advances on Nuclear Genomes of Economically Important Trees of Lauraceae [J]. Chinese Bulletin of Botany, 2024, 59(2): 302-318. |
| [14] | Chen-Kun Jiang, Wen-Bin Yu, Guang-Yuan Rao, Huaicheng Li, Julien B. Bachelier, Hartmut H. Hilger, Theodor C. H. Cole. Plant Phylogeny Posters—An educational project on plant diversity from an evolutionary perspective [J]. Biodiv Sci, 2024, 32(11): 24210-. |
| [15] | Yi Song, Hanghang Chen, Xin Cui, Zhifeng Lu, Shipeng Liao, Yangyang Zhang, Xiaokun Li, Rihuan Cong, Tao Ren, Jianwei Lu. Potassium Nutrient Status-mediated Leaf Growth of Oilseed Rape (Brassica napus) and Its Effect on Phyllosphere Microorganism [J]. Chinese Bulletin of Botany, 2024, 59(1): 54-65. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||