

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (5): 620-630.DOI: 10.11983/CBB16006 cstr: 32102.14.CBB16006
• Orginal Article • Previous Articles Next Articles
Jia Ledong, Li Shimeng, Xu Daixiang, Qu Cunmin, Li Jiana, Wang Rui*( )
)
Received:2016-01-11
Accepted:2016-04-26
Online:2016-09-01
Published:2018-08-10
Contact:
Wang Rui
About author:# Co-first authors
Jia Ledong, Li Shimeng, Xu Daixiang, Qu Cunmin, Li Jiana, Wang Rui. Bioinformatics Analysis of BnMYB80 Genes in Brassica napus [J]. Chinese Bulletin of Botany, 2016, 51(5): 620-630.
| Gene name | Gene ID | Chromosome | Length (aa) | MW (kDa) | pI | Instability | Aliphatic index | Gravy |
|---|---|---|---|---|---|---|---|---|
| BrMYB80A02 | 103851848 | A02 | 320 | 36.1054 | 6.95 | 35.89 | 65.88 | -0.808 |
| BrMYB80A10 | 103845046 | A10 | 320 | 35.8452 | 7.24 | 33.15 | 69.81 | -0.721 |
| BolMYB80C02 | 106327859 | C02 | 324 | 36.5430 | 7.75 | 37.04 | 66.57 | -0.790 |
| BolMYB80C09 | 106317148 | C09 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80A02 | 106389601 | A02 | 322 | 36.3678 | 6.95 | 35.55 | 65.47 | -0.792 |
| BnMYB80A10 | 674915774 | A10 | 320 | 35.8232 | 7.24 | 34.62 | 68.59 | -0.712 |
| BnMYB80C02 | 106379735 | C02 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80C03 | 106385056 | C03 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80C05 | 106400365 | C05 | 319 | 35.7842 | 6.90 | 33.76 | 68.50 | -0.738 |
| BnMYB80Cnn | 106389601 | Cnn | 324 | 36.5490 | 6.93 | 35.65 | 66.57 | -0.771 |
Table 1 Physicochemical parameters of MYB80 transcription factors
| Gene name | Gene ID | Chromosome | Length (aa) | MW (kDa) | pI | Instability | Aliphatic index | Gravy |
|---|---|---|---|---|---|---|---|---|
| BrMYB80A02 | 103851848 | A02 | 320 | 36.1054 | 6.95 | 35.89 | 65.88 | -0.808 |
| BrMYB80A10 | 103845046 | A10 | 320 | 35.8452 | 7.24 | 33.15 | 69.81 | -0.721 |
| BolMYB80C02 | 106327859 | C02 | 324 | 36.5430 | 7.75 | 37.04 | 66.57 | -0.790 |
| BolMYB80C09 | 106317148 | C09 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80A02 | 106389601 | A02 | 322 | 36.3678 | 6.95 | 35.55 | 65.47 | -0.792 |
| BnMYB80A10 | 674915774 | A10 | 320 | 35.8232 | 7.24 | 34.62 | 68.59 | -0.712 |
| BnMYB80C02 | 106379735 | C02 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80C03 | 106385056 | C03 | 320 | 35.9113 | 7.22 | 31.94 | 68.56 | -0.740 |
| BnMYB80C05 | 106400365 | C05 | 319 | 35.7842 | 6.90 | 33.76 | 68.50 | -0.738 |
| BnMYB80Cnn | 106389601 | Cnn | 324 | 36.5490 | 6.93 | 35.65 | 66.57 | -0.771 |
| Gene name | CDS (bp) | Exon | MYB80 conserved domain (aa) | ||||
|---|---|---|---|---|---|---|---|
| SANT | SANT | IRO | KNOX1 | Cir_N | |||
| BrMYB80A02 | 1772 | 3 | 13-63 | 66-114 | - | - | - |
| BrMYB80A10 | 1631 | 3 | 13-63 | 66-114 | - | - | - |
| BolMYB80C02 | 1891 | 4 | 13-63 | 66-114 | 123-140 | 130-173 | 246-282 |
| BolMYB80C09 | 1573 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80A02 | 1653 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | 244-280 |
| BnMYB80A10 | 1471 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80C02 | 1471 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80C03 | 1471 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | - |
| BnMYB80C05 | 1461 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | - |
| BnMYB80Cnn | 1668 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | 246-282 |
Table 2 Analysis of conserved domains of MYB80 transcription factors
| Gene name | CDS (bp) | Exon | MYB80 conserved domain (aa) | ||||
|---|---|---|---|---|---|---|---|
| SANT | SANT | IRO | KNOX1 | Cir_N | |||
| BrMYB80A02 | 1772 | 3 | 13-63 | 66-114 | - | - | - |
| BrMYB80A10 | 1631 | 3 | 13-63 | 66-114 | - | - | - |
| BolMYB80C02 | 1891 | 4 | 13-63 | 66-114 | 123-140 | 130-173 | 246-282 |
| BolMYB80C09 | 1573 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80A02 | 1653 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | 244-280 |
| BnMYB80A10 | 1471 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80C02 | 1471 | 3 | 13-63 | 66-114 | 123-140 | 130-173 | - |
| BnMYB80C03 | 1471 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | - |
| BnMYB80C05 | 1461 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | - |
| BnMYB80Cnn | 1668 | 3 | 13-63 | 66-144 | 123-140 | 130-173 | 246-282 |
| Gene name | α helix (%) | Extend strand (%) | β turn (%) | Random coil (%) |
|---|---|---|---|---|
| BrMYB80A02 | 41.25 | 11.56 | 10.00 | 37.19 |
| BrMYB80A10 | 37.81 | 14.06 | 10.00 | 38.12 |
| BolMYB80C02 | 44.14 | 10.19 | 8.64 | 37.04 |
| BolMYB80C09 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80A02 | 42.55 | 11.18 | 9.63 | 36.65 |
| BnMYB80A10 | 37.19 | 15.00 | 10.31 | 37.50 |
| BnMYB80C02 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80C03 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80C05 | 36.68 | 14.73 | 10.03 | 38.56 |
| BnMYB80Cnn | 44.14 | 10.49 | 9.57 | 35.80 |
Table 3 Predicted secondary structure of MYB80 protein
| Gene name | α helix (%) | Extend strand (%) | β turn (%) | Random coil (%) |
|---|---|---|---|---|
| BrMYB80A02 | 41.25 | 11.56 | 10.00 | 37.19 |
| BrMYB80A10 | 37.81 | 14.06 | 10.00 | 38.12 |
| BolMYB80C02 | 44.14 | 10.19 | 8.64 | 37.04 |
| BolMYB80C09 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80A02 | 42.55 | 11.18 | 9.63 | 36.65 |
| BnMYB80A10 | 37.19 | 15.00 | 10.31 | 37.50 |
| BnMYB80C02 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80C03 | 35.94 | 15.31 | 10.94 | 37.81 |
| BnMYB80C05 | 36.68 | 14.73 | 10.03 | 38.56 |
| BnMYB80Cnn | 44.14 | 10.49 | 9.57 | 35.80 |
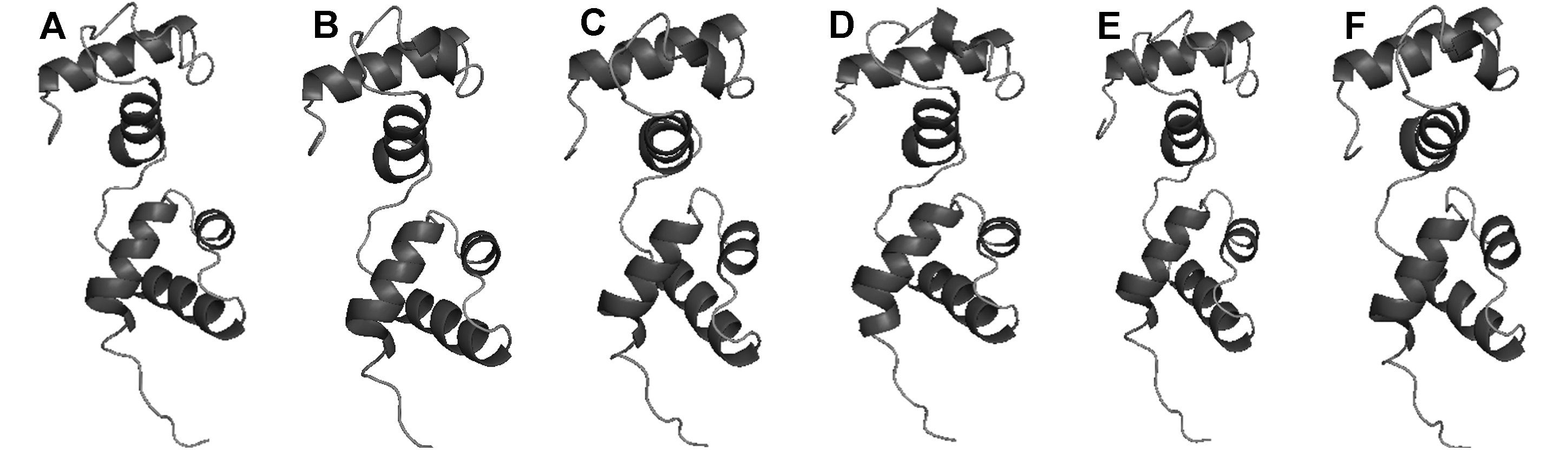
Figure 2 Predicted tertiary structure of BnMYB80 proteins (A) BnMYB80A02; (B) BnMYB80A10; (C) BnMYB80C02; (D) BnMYB80C03; (E) BnMYB80C05; (F) BnMYB80Cnn
| 1 | 陈凤祥, 胡宝成, 李强生, 张曼琳 (1993). 甘蓝型油菜细胞核雄性不育材料9012A的发现与初步研究. 北京农业大学学报 9, 57-61. |
| 2 | 邓志刚, 金樑, 李晶, 王文斌, 杨龙, 王晓娟 (2013). MYB类转录因子对花药和花粉发育的调控途径. 西北植物学报 33, 850-856. |
| 3 | 方子君, 石其龙, 杨仲南, 张森 (2008). 水稻OsMS2基因在花药发育中的功能分析. 植物学通报 25, 665-672. |
| 4 | 郭弘光, 吴繁花 (2012). MYB转录因子功能与调控研究进展. 安徽农业科学 40, 10381-10383,10516. |
| 5 | 侯国佐, 王华, 张瑞茂 (1990). 甘蓝型油菜细胞核雄性不育材料117A的遗传研究. 中国油料 2, 7-10. |
| 6 | 胡胜武, 于澄宇, 赵惠贤, 路明 (2002). 甘蓝型油菜显性核不育材料Shaan-GMS纯合两型系803AB的选育. 西北农业学报 11, 25-27. |
| 7 | 李树林, 周志疆, 周熙荣 (1993). 甘蓝型油菜隐性核不育系S45AB的遗传. 上海农业学报 9, 1-7. |
| 8 | 刘琪迩, 杜坤, 王幼平 (2015). 油菜细胞质雄性不育与育性恢复机理的研究进展. 生物技术通报 31, 15-22. |
| 9 | 刘淑娟, 朱祺, 幸学俊, 王碧琴, 余发新, 周华 (2014). 植物雄性不育影响因素研究进展. 中国农学通报 30, 46-50. |
| 10 | 宋来强, 傅廷栋, 杨光圣, 涂金星, 马朝芝 (2005). 1对复等位基因控制的油菜(Brassica napus)显性核不育系609AB的遗传验证. 作物学报 31, 869-875. |
| 11 | 孙超才, 赵华, 王伟荣, 李延莉, 钱小芳, 方光华 (2002). 甘蓝型油菜隐性核不育系20118A的遗传与利用探讨. 中国油料作物学报 24, 1-4. |
| 12 | 王超, 安学丽, 张增为, 杨青, 饶力群, 陈信波, 方才臣, 万向元 (2013). 植物隐性核雄性不育基因育种技术体系的研究进展与展望. 中国生物工程杂志 33, 124-130. |
| 13 | 王瑞, 李加纳, 唐章林, 谌利, 徐新福 (2005). 显性核不育油菜圆叶纯合两型系 GdlAB的选育. 中国农学通报 21, 174-175, 229. |
| 14 | 王通强, 田筑萍, 黄泽素, 魏忠芬, 邵明波 (1999). 甘蓝型双低油菜细胞核显性雄性不育系黔油2AB的选育. 贵州农业科学 27, 14-18. |
| 15 | 周熙荣, 庄静, 孙超才, 王伟荣, 李延莉, 杨立勇, 蒋美艳, 顾龙弟, 钱小芳 (2010). 甘蓝型油菜显性核不育纯合两型系“HY15AB”的选育. 上海农业学报 26, 1-4. |
| 16 | 俎峰, 夏胜前, 顿小玲, 周正富, 曾芳琴, 易斌, 文静, 马朝芝, 沈金雄, 涂金星, 傅廷栋 (2010). 基于分子标记的油菜隐性核不育7-7365AB遗传模式探究. 中国农业科学 43, 3067-3075. |
| 17 | Cai CF, Zhu J, Lou Y, Guo ZL, Xiong SX, Wang K, Yang ZN (2015). The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development bet- ween rice and Arabidopsis.Sci Bull 60, 1073-1082. |
| 18 | Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Silva CD, Just J, Falentin C, Koh CS, Clainche IL, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Paslier MCL, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950-953. |
| 19 | Feng BM, Lu DH, Ma X, Peng YB, Sun YJ, Ning G, Ma H (2012). Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development.Plant J 72, 612-624. |
| 20 | Graybosch RA, Bernard RL, Cremeens CR, Palmer RG (1984). Genetic and cytological studies of a male-sterile, female-fertile soybean mutant, a new male-sterile gene (ms2) in Glycine max (L.) Merr.J Hered 75, 383-388. |
| 21 | Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R (1993). The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1, 3-glucanases.Plant J 4, 1023-1033. |
| 22 | Jiang LX, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005). VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17, 584-596. |
| 23 | Li SF, Higginson T, Parish RW (1999). A novel MYB- related gene from Arabidopsis thaliana expressed in developing anthers.Plant Cell Physiol 40, 343-347. |
| 24 | Li SF, Iacuone S, Parish RW (2007). Suppression and restoration of male fertility using a transcription factor. Plant Biotechnol J 5, 297-312. |
| 25 | Liu SY, Liu YM, Yang XH, Tong CB, Edwards D, Parkin IAP, Zhao MX, Ma JX, Yu JY, Huang SM, Wang XY, Wang JY, Lu K, Fang ZY, Bancroft I, Yang TJ, Hu Q, Wang XF, Yue Z, Li HJ, Yang LF, Wu J, Zhou Q, Wang WX, King GJ, Pires JC, Lu CX, Wu ZY, Sampath P, Wang Z, Guo H, Pan SK, Yang LM, Min JM, Zhang D, Jin DC, Li WS, Belcram H, Tu JX, Guan M, Qi CQ, Du DZ, Li JN, Jiang LC, Batley J, Sharpe AG, Park BS, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong CH, Wang L, Li JP, Hu ZY, Zhuang M, Huang Y, Huang JY, Shi JQ, Mei DS, Liu J, Lee TH, Wang JP, Jin HZ, Li ZY, Li X, Zhang JF, Xiao L, Zhou YM, Liu ZS, Liu XQ, Qin R, Tang X, Liu WB, Wang YP, Zhang YY, Lee J, Kim HH, Denoeud F, Xu X, Liang XM, Hua W, Wang XW, Wang J, Chalhoub B, Paterson AH (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes.Nat Commun 5, 3930. |
| 26 | Lu PL, Chai MF, Yang JG, Ning G, Wang GL, Ma H (2014). The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis.Plant Physiol 164, 1893-1904. |
| 27 | McCormick S (2004). Control of male gametophyte develop- ment.Plant Cell 16, 142-153. |
| 28 | Pabo CO, Sauer RT (1992). Transcription factors: structural families and principles of DNA recognition.Annu Rev Biochem 61, 1053-1095. |
| 29 | Parish RW, Li SF (2010). Death of a tapetum: a programme of development altruism.Plant Sci 178, 73-89. |
| 30 | Phan HA, Iacuone S, Li SF, Parish RW (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23, 2209-2224. |
| 31 | Phan HA, Li SF, Parish RW (2012). MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol Biol 78, 171-183. |
| 32 | Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor.Plant J 33, 413-423. |
| 33 | Wang XW, Wang HZ, Wang J, Sun RF, Wu J, Liu SY, Bai YQ, Mun JH, Bancroft I, Cheng F, Huang SW, Li XX, Hua W, Wang JY, Wang XY, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu BH, Li B, Liu B, Tong CB, Song C, Duran C, Peng CF, Geng CY, Koh C, Lin CY, Edwards D, Mu DS, Shen D, Soumpourou E, Li F, Fraser F, Lassalle G, King GJ, Bonnema G, Tang HB, Wang HP, Belcram H, Zhou HL, Hirakawa H, Abe H, Guo H, Wang H, Jin HZ, Parkin IAP, Batley J, Kim JS, Just J, Li JW, Xu JH, Deng J, Kim JA, Li JP, Yu JY, Meng JL, Wang JP, Min JM, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao MX, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai QL, Huang QF, Li RQ, Tabata S, Cheng SF, Zhang S, Zhang SJ, Huang SM, Sato S, Sun SL, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li YR, Du YC, Liao YC, Lim Y, Narusaka Y, Wang YP, Wang ZY, Li ZY, Wang ZW, Xiong ZY, Zhang ZH (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43, 1035-1039. |
| 34 | Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001). The Arabidopsis MALE STERILITY (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28, 27-39. |
| 35 | Xu Y, Lacuone S, Li SF, Parish RW (2014). MYB80 homo- logues in Arabidopsis, cotton and Brassica: regulation and functional conservation in tapetal and pollen development.BMC Plant Biol 14, 278. |
| 36 | Zhang S, Fang ZJ, Zhu J, Gao JF, Yang ZN (2010). OsMYB103 is required for rice anther development by regulating tapetum development and exine formation.Sci Bull 55, 3288-3297. |
| 37 | Zhang W, Sun Y, Timofejeva L, Chen CB, Grossniklaus U, Ma H (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPET- UM1 (DYT1) encoding a putative bHLH transcription factor.Development 133, 3085-3095. |
| 38 | Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis.Plant J 52, 528-538. |
| 39 | Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008). Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis.Plant J 55, 266-277. |
| [1] | Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli. Identification of the Spinach AT-hook Gene Family and Analysis of Expression Profiles [J]. Chinese Bulletin of Botany, 2025, 60(3): 377-392. |
| [2] | Liuqing Yang, Jin Wang, Jingli Yan, Qinqin Chen, Haokun Cheng, Chun Li, Peiyu Zhao, Bo Yang, Yuanqing Jiang. Analysis of Expression Characteristics and Identification of Interaction Proteins of BnaABF2 Transcription Factor in Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 49-61. |
| [3] | Qingyang Li, Cui Liu, Li He, Shan Peng, Jiayin Ma, Ziyi Hu, Hongbo Liu. Cloning and Functional Analysis of the BnaA02.CPSF6 Gene from Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 62-73. |
| [4] | Yi Song, Hanghang Chen, Xin Cui, Zhifeng Lu, Shipeng Liao, Yangyang Zhang, Xiaokun Li, Rihuan Cong, Tao Ren, Jianwei Lu. Potassium Nutrient Status-mediated Leaf Growth of Oilseed Rape (Brassica napus) and Its Effect on Phyllosphere Microorganism [J]. Chinese Bulletin of Botany, 2024, 59(1): 54-65. |
| [5] | Zhang Yingchuan, Wu Xiaomingyu, Tao Baolong, Chen Li, Lu Haiqin, Zhao Lun, Wen Jing, Yi Bin, Tu Jinxing, Fu Tingdong, Shen Jinxiong. Bna-miR43 Mediates the Response of Drought Tolerance in Brassica napus [J]. Chinese Bulletin of Botany, 2023, 58(5): 701-711. |
| [6] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [7] | Cong Xu, Feiyu Zhang, Daoyuan Yu, Xin Sun, Feng Zhang. Performance evaluation of molecular taxonomy assignment tools for soil invertebrates [J]. Biodiv Sci, 2022, 30(12): 22252-. |
| [8] | Yuhui Zhao, Xiuxiu Li, Zhuo Chen, Hongwei Lu, Yucheng Liu, Zhifang Zhang, Chengzhi Liang. An Overview of Genome-wide Association Studies in Plants [J]. Chinese Bulletin of Botany, 2020, 55(6): 715-732. |
| [9] | Zeyuan Zuo,Wanlin Liu,Jie Xu. Evolution and Functional Analysis of Gene Clusters in Anther Tapetum Cells of Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2020, 55(2): 147-162. |
| [10] | Min Song,Yao Zhang,Liying Wang,Xiangyong Peng. Genome-wide Identification and Phylogenetic Analysis of Zinc Finger Homeodomain Family Genes in Brassica napus [J]. Chinese Bulletin of Botany, 2019, 54(6): 699-710. |
| [11] | Liu Wei, Tong Yong’ao, Bai Jie. Bioinformatics Analysis of tRNA-derived Fragments in Rice Male Gametophyte Development [J]. Chinese Bulletin of Botany, 2018, 53(5): 625-633. |
| [12] | Liu Kaige, Qi Shuanghui, Duan Shaowei, Li Dong, Jin Changyu, Gao Chenhao, Liu Mingxun Chen Xuanxia. Functional Analysis of Brassica napus BnTTG1-1 Gene [J]. Chinese Bulletin of Botany, 2017, 52(6): 713-722. |
| [13] | Gao Huhu, Zhang Yunxiao, Hu Shengwu, Guo Yuan. Genome-wide Survey and Phylogenetic Analysis of MADS-box Gene Family in Brassica napus [J]. Chinese Bulletin of Botany, 2017, 52(6): 699-712. |
| [14] | Tian Cheng, Qiang Wei, Guanglin Li. Bioinformatics Analysis of the TPS Gene Family in Coffee canephora [J]. Chinese Bulletin of Botany, 2016, 51(2): 235-250. |
| [15] | Xiaoting Xu, Zhiheng Wang, Dimitar Dimitrov, Carsten Rahbek. Using NCBIminer to search and download nucleotide sequences from GenBank [J]. Biodiv Sci, 2015, 23(4): 550-555. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||