

植物学报 ›› 2025, Vol. 60 ›› Issue (5): 669-678.DOI: 10.11983/CBB25052 cstr: 32102.14.CBB25052
• 热点评述 • 下一篇
收稿日期:2025-04-01
接受日期:2025-06-03
出版日期:2025-09-10
发布日期:2025-06-10
通讯作者:
*E-mail: yuening94@163.com
作者简介:刘玉乐, 清华大学生命科学学院教授, 国家杰出青年基金获得者。长期从事植物病毒和细胞自噬相关研究, 在植物抗病毒免疫、病毒病理、病毒载体及细胞自噬方面做出了一些有国际影响的工作。以通讯作者或第一作者在Nature、Cell和Cell Host & Microbe等期刊发表论文60余篇。主编教材《生物化学》(清华大学出版社, 2023)。论文被Web of Science共引用超过21829次。自2014年起每年均入选Elsevier发布的中国高被引学者榜单
基金资助:
Liu Deshui1, Yue Ning2,*( ), Liu Yule2
), Liu Yule2
Received:2025-04-01
Accepted:2025-06-03
Online:2025-09-10
Published:2025-06-10
Contact:
*E-mail: yuening94@163.com
摘要: 近年来, 植物抗病免疫研究取得了突破性进展, 包括病原识别、免疫信号转导及植物-病原-介体-环境互作等。这些研究不仅增强了我们对植物抗病免疫的理解, 还为分子育种和分子遗传学研究奠定了坚实的基础。近期, 国内多家单位相继在植物免疫机制研究中取得了令人振奋的新突破, 从植物应对病原的识别机制、次级代谢产物参与植物抗病反应过程、 禾本科作物的抗病模块和基于人工智能的抗病小肽设计等不同层面对植物免疫反应的分子机制进行了深入解析。随着CRISPR/Cas9基因编辑技术和人工智能的快速发展, 这些研究成果将有助于创制具有抗病特性的新种质, 从而加速抗病作物新品种的培育过程, 对于抗病生物育种和国家粮食安全具有重要意义。
刘德水, 岳宁, 刘玉乐. 植物免疫机制新突破. 植物学报, 2025, 60(5): 669-678.
Liu Deshui, Yue Ning, Liu Yule. Emerging Innovation in Plant Immunity. Chinese Bulletin of Botany, 2025, 60(5): 669-678.
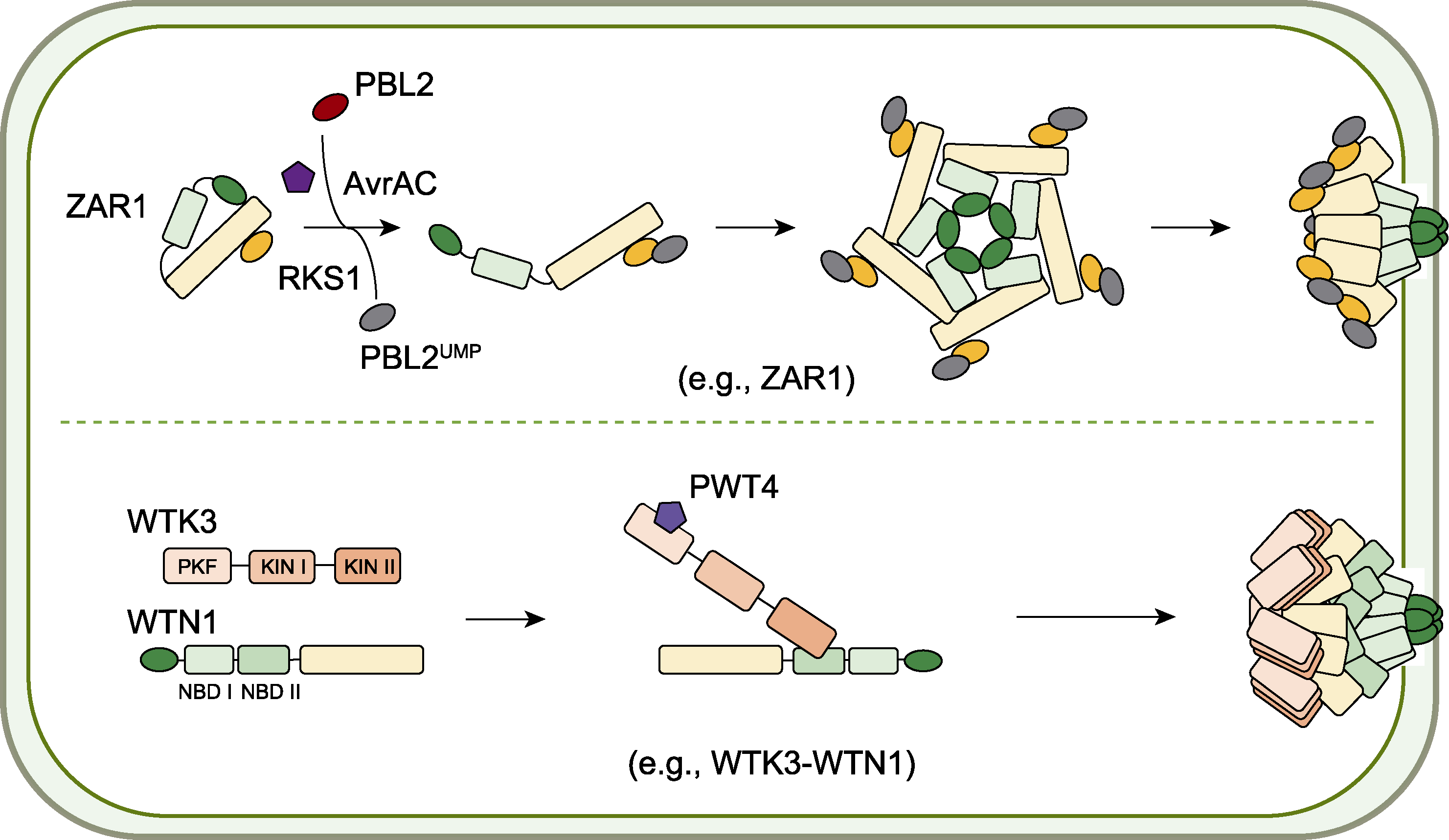
图1 抗病小体的组装机制 上半部分显示拟南芥ZAR1形成抗病小体的过程。ZAR1与类受体激酶RKS1在细胞质中互作, 识别被效应因子AvrAC尿苷化修饰的PBL2后, 其构象发生改变, 形成抗病小体发挥钙离子通道作用。下半部分显示小麦WTN1形成抗病小体的过程。WTK3的PKF结构域与效应因子PWT4结合后促进WTN1多聚, 形成钙渗透性离子通道。
Figure 1 Resistosome assembly mechanisms Top panel: Arabidopsis ZAR1 resistosome formation. ZAR1 interacts with the receptor-like kinase RKS1 in cytoplasm. Upon recognition of PBL2 uridylylated by the effector AvrAC, ZAR1 undergoes conformational changes to assemble into a resistosome that functions as a calcium channel. Bottom panel: Wheat WTN1 resistosome assembly. Binding of the effector PWT4 to the pseudo-kinase fragment (PKF) domain of WTK3 promotes oligomerization of WTN1, forming a calcium-permeable ion channel.
| [1] | Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O'Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Žemgulytė A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Žídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493-500. |
| [2] |
Ahuja I, Kissen R, Bones AM (2012). Phytoalexins in defense against pathogens. Trends Plant Sci 17, 73-90.
DOI PMID |
| [3] |
Bednarek P (2012). Chemical warfare or modulators of defence responses—the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15, 407-414.
DOI PMID |
| [4] |
Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze- Lefert P (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101-106.
DOI PMID |
| [5] |
Bi GZ, Su M, Li N, Liang Y, Dang S, Xu JC, Hu MJ, Wang JZ, Zou MX, Deng YN, Li QY, Huang SJ, Li JJ, Chai JJ, He KM, Chen YH, Zhou JM (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528-3541.
DOI PMID |
| [6] | Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D'Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo NX, Luo MC, Sehgal S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N (2012). Analysis of the bread wheat genome using whole- genome shotgun sequencing. Nature 491, 705-710. |
| [7] | Chen RJ, Chen J, Powell OR, Outram MA, Arndell T, Gajendiran K, Wang YL, Lubega J, Xu Y, Ayliffe MA, Blundell C, Figueroa M, Sperschneider J, Vanhercke T, Kanyuka K, Tang DZ, Zhong GT, Gardener C, Yu GT, Gourdoupis S, Jaremko Ł, Matny O, Steffenson BJ, Boshoff WHP, Meyer WB, Arold ST, Dodds PN, Wulff BBH (2025). A wheat tandem kinase activates an NLR to trigger immunity. Science 387, 1402-1408. |
| [8] | Chen SS, Rouse MN, Zhang WJ, Zhang XQ, Guo Y, Briggs J, Dubcovsky J (2020). Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol 225, 948-959. |
| [9] | Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Ara- bidopsis. Science 331, 1185-1188. |
| [10] |
Feng F, Zhou JM (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15, 469-476.
DOI PMID |
| [11] | Förderer A, Li ET, Lawson AW, Deng YN, Sun Y, Logemann E, Zhang XX, Wen J, Han ZF, Chang JB, Chen YH, Schulze-Lefert P, Chai JJ (2022). A wheat resistosome defines common principles of immune receptor channels. Nature 610, 532-539. |
| [12] | Graham JH, Bassanezi RB, Dawson WO, Dantzler R (2024). Management of Huanglongbing of citrus: lessons from São Paulo and Florida. Ann Rev Phytopathol 62, 243-262. |
| [13] |
Halkier BA, Gershenzon J (2006). Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57, 303-333.
PMID |
| [14] |
Huang SJ, Jia AL, Ma SC, Sun Y, Chang XY, Han ZF, Chai JJ (2023). NLR signaling in plants: from resistosomes to second messengers. Trends Biochem Sci 48, 776-787.
DOI PMID |
| [15] | Huang Y, Yang JL, Sun X, Li JH, Cao XQ, Yao SZ, Han YH, Chen CT, Du LL, Li S, Ji YH, Zhou T, Wang H, Han JJ, Wang WM, Wei CH, Xie Q, Yang ZR, Li Y (2025). Perception of viral infections and initiation of antiviral defence in rice. Nature 641, 173-181. |
| [16] | Jones JDG, Dangl JL (2006). The plant immune system. Nature 444, 323-329. |
| [17] |
Joubert A, Simoneau P, Campion C, Bataillé-Simoneau N, Iacomi-Vasilescu B, Poupard P, François JM, Georgeault S, Sellier E, Guillemette T (2011). Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol Microbiol 79, 1305-1324.
DOI PMID |
| [18] | Klymiuk V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen SS, Feng LH, Frenkel Z, Krugman T, Lidzbarsky G, Chang W, Jääskeläinen MJ, Schudoma C, Paulin L, Laine P, Bariana H, Sela H, Saleem K, Sørensen CK, Hovmøller MS, Distelfeld A, Chalhoub B, Dubcovsky J, Korol AB, Schulman AH, Fahima T (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun 9, 3735. |
| [19] | Li SN, Lin DX, Zhang YW, Deng M, Chen YX, Lv B, Li BS, Lei Y, Wang YP, Zhao L, Liang YT, Liu JX, Chen KL, Liu ZY, Xiao J, Qiu JL, Gao CX (2022a). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455-460. |
| [20] | Li ZL, Yang XX, Li WL, Wen ZY, Duan JN, Jiang ZH, Zhang DL, Xie XL, Wang XT, Li FF, Li DW, Zhang YL (2022b). SAMDC3 enhances resistance to Barley stripe mosaic virus by promoting the ubiquitination and proteasomal degradation of viral γb protein. New Phytol 234, 618-633. |
| [21] |
Lu P, Guo L, Wang ZZ, Li BB, Li J, Li YH, Qiu D, Shi WQ, Yang LJ, Wang N, Guo GH, Xie JZ, Wu QH, Chen YX, Li MM, Zhang HZ, Dong LL, Zhang PP, Zhu KY, Yu DZ, Zhang Y, Deal KR, Huo NX, Liu CM, Luo MC, Dvorak J, Gu YQ, Li HJ, Liu ZY (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun 11, 680.
DOI PMID |
| [22] | Lu P, Zhang GH, Li J, Gong Z, Wang GJ, Dong LL, Zhang HZ, Guo GH, Su M, Wang K, Wang YM, Zhu KY, Wu QH, Chen YX, Li MM, Huang BG, Li BB, Li WL, Dong L, Hou YK, Cui XJ, Fu HK, Qiu D, Yuan CG, Li HJ, Zhou JM, Han GZ, Chen YH, Liu ZY (2025). A wheat tandem kinase and NLR pair confers resistance to multiple fungal pathogens. Science 387, 1418-1424. |
| [23] | Miao P, Wang HJ, Wang W, Wang ZD, Ke H, Cheng HY, Ni JJ, Liang JN, Yao YF, Wang JZ, Zhou JM, Lei XG (2025). A widespread plant defense compound disarms bacterial type III injectisome assembly. Science 387, eads0377. |
| [24] |
Mohanty SP, Hughes DP, Salathé M (2016). Using deep learning for image-based plant disease detection. Front Plant Sci 7, 1419.
PMID |
| [25] | Ngou BPM, Ahn HK, Ding PT, Jones JDG (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110-115. |
| [26] | Ofer D, Brandes N, Linial M (2021). The language of proteins: NLP, machine learning & protein sequences. Comput Struct Biotechnol J 19, 1750-1758. |
| [27] | Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, García-Casado G, Witters E, Inzé D, Long JA, De Jaeger G, Solano R, Goossens A (2010). NINJA connects the co-repressor TOPLESS to jasmonate signaling. Nature 464, 788-791. |
| [28] |
Piasecka A, Jedrzejczak-Rey N, Bednarek P (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206, 948-964.
DOI PMID |
| [29] |
Reveguk T, Fatiukha A, Potapenko E, Reveguk I, Sela H, Klymiuk V, Li YH, Pozniak C, Wicker T, Coaker G, Fahima T (2025). Tandem kinase proteins across the plant kingdom. Nat Genet 57, 254-262.
DOI PMID |
| [30] | Rogers EE, Glazebrook J, Ausubel FM (1996). Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact 9, 748-757. |
| [31] | Sellam A, Dongo A, Guillemette T, Hudhomme P, Simoneau P (2007). Transcriptional responses to exposure to the brassicaceous defence metabolites camalexin and allyl-isothiocyanate in the necrotrophic fungus Alternaria brassicicola. Mol Plant Pathol 8, 195-208. |
| [32] | Sheard LB, Tan X, Mao HB, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, He SY, Rizo J, Howe GA, Zheng N (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co- receptor. Nature 468, 400-405. |
| [33] |
Shen QT, Hu T, Bao M, Cao LG, Zhang HW, Song FM, Xie Q, Zhou XP (2016). Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol Plant 9, 911-925.
DOI PMID |
| [34] | Shlezinger N, Minz A, Gur Y, Hatam I, Dagdas YF, Talbot NJ, Sharon A (2011). Anti-apoptotic machinery protects the necrotrophic fungus Botrytis cinerea from host-induced apoptotic-like cell death during plant infection. PLoS Pathog 7, e1002185. |
| [35] | Tang XY, Xiao YM, Zhou JM (2006). Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact 19, 1159-1166. |
| [36] | Tian HN, Wu ZS, Chen SY, Ao K, Huang WJ, Yaghmaiean H, Sun TJ, Xu F, Zhang YJ, Wang SC, Li X, Zhang YL (2021). Activation of TIR signaling boosts pattern-triggered immunity. Nature 598, 500-503. |
| [37] | Vargas P, Farias GA, Nogales J, Prada H, Carvajal V, Barón M, Rivilla R, Martín M, Olmedilla A, Gallegos MT (2013). Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ Microbiol Rep 5, 841-850. |
| [38] | Vargas P, Felipe A, Michán C, Gallegos MT (2011). Induction of Pseudomonas syringae pv. tomato DC3000 MexAB-OprM multidrug efflux pump by flavonoids is mediated by the repressor PmeR. Mol Plant Microbe Interact 24, 1207-1219. |
| [39] | Wang JZ, Hu MJ, Wang J, Qi JF, Han ZF, Wang GX, Qi YJ, Wang HW, Zhou JM, Chai JJ (2019a). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870. |
| [40] | Wang JZ, Wang J, Hu MJ, Wu S, Qi JF, Wang GX, Han ZF, Qi YJ, Gao N, Wang HW, Zhou JM, Chai JJ (2019b). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868. |
| [41] |
Wang W, Yang J, Zhang J, Liu YX, Tian CP, Qu BY, Gao CL, Xin PY, Cheng SJ, Zhang WJ, Miao P, Li L, Zhang XJ, Chu JF, Zuo JR, Li JY, Bai Y, Lei XG, Zhou JM (2020). An Arabidopsis secondary metabolite directly targets expression of the bacterial type III secretion system to inhibit bacterial virulence. Cell Host Microbe 27, 601-613.
DOI PMID |
| [42] |
Wang Y, Pruitt RN, Nürnberger T, Wang YC (2022). Evasion of plant immunity by microbial pathogens. Nat Rev Microbiol 20, 449-464.
DOI PMID |
| [43] |
Wang YJ, Gong Q, Wu YY, Huang F, Ismayil A, Zhang DF, Li HG, Gu HQ, Ludman M, Fátyol K, Qi YJ, Yoshioka K, Hanley-Bowdoin L, Hong YG, Liu YL (2021). A calmodulin-binding transcription factor links calcium signaling to antiviral RNAi defense in plants. Cell Host Microbe 29, 1393-1406.
DOI PMID |
| [44] |
Washburn JD, Mejia-Guerra MK, Ramstein G, Kremling KA, Valluru R, Buckler ES, Wang H (2019). Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence. Proc Natl Acad Sci USA 116, 5542-5549.
DOI PMID |
| [45] | Wu DW, Qi TC, Li WX, Tian HX, Gao H, Wang JJ, Ge J, Yao RF, Ren CM, Wang XB, Liu YL, Kang L, Ding SW, Xie DX (2017a). Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res 27, 402-415. |
| [46] | Wu JG, Yang RX, Yang ZR, Yao SZ, Zhao SS, Wang Y, Li PC, Song XW, Jin L, Zhou T, Lan Y, Xie LH, Zhou XP, Chu CC, Qi YJ, Cao XF, Li Y (2017b). ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants 3, 16203. |
| [47] | Wu JG, Yang ZR, Wang Y, Zheng LJ, Ye RQ, Ji YH, Zhao SS, Ji SY, Liu RF, Xu L, Zheng H, Zhou YJ, Zhang X, Cao XF, Xie LH, Wu ZJ, Qi YJ, Li Y (2015). Viral-inducible argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4, e05733. |
| [48] | Wu JG, Zhang YL, Li FF, Zhang XM, Ye J, Wei TY, Li ZH, Tao XR, Cui F, Wang XB, Zhang LL, Yan F, Li SF, Liu YL, Li DW, Zhou XP, Li Y (2024). Plant virology in the 21st century in China: recent advances and future directions. J Integr Plant Biol 66, 579-622. |
| [49] | Xin XF, Kvitko B, He SY (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16, 316-328. |
| [50] |
Yang ZR, Huang Y, Yang JL, Yao SZ, Zhao K, Wang DH, Qin QQ, Bian Z, Li Y, Lan Y, Zhou T, Wang H, Liu C, Wang WM, Qi YJ, Xu ZH, Li Y (2020). Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89-103.
DOI PMID |
| [51] | Yuan MH, Jiang ZY, Bi GZ, Nomura K, Liu MH, Wang YP, Cai BY, Zhou JM, He SY, Xin XF (2021). Pattern- recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105-109. |
| [52] | Zhao PZ, Yang H, Sun YW, Zhang JY, Gao KX, Wu JB, Zhu CR, Yin CC, Chen XY, Liu Q, Xia QD, Li Q, Xiao H, Sun HX, Zhang XX, Yi L, Zhou CY, Kliebenstein DJ, Fang RX, Wang XF, Ye J (2025). Targeted MYC2 stabilization confers citrus Huanglongbing resistance. Science 388, 191-198. |
| [53] | Zhou JM, Zhang YL (2020). Plant immunity: danger perception and signaling. Cell 181, 978-989. |
| [54] |
Zhou WK, Lozano-Torres JL, Blilou I, Zhang XY, Zhai QZ, Smant G, Li CY, Scheres B (2019). A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942-956.
DOI PMID |
| [1] | 肖银燕, 于华, 万里. 植物免疫研究: 机制突破和应用创新[J]. 植物学报, 2025, 60(5): 693-703. |
| [2] | 徐羽丰, 周冕. 植物免疫的转录后调控[J]. 植物学报, 2025, 60(5): 704-721. |
| [3] | 王笑, 徐昌文, 钱虹萍, 李思博, 林金星, 崔亚宁. 植物细胞壁参与免疫反应的机制及其原位非标记成像方法[J]. 植物学报, 2025, 60(5): 773-785. |
| [4] | 江亚楠, 徐雨青, 魏毅铤, 陈钧, 张容菀, 赵蓓蓓, 林宇翔, 饶玉春. 水稻抗病调控机制研究进展[J]. 植物学报, 2025, 60(5): 734-748. |
| [5] | 覃磊, 彭志红, 夏石头. 植物NLR免疫受体的识别、免疫激活与信号调控[J]. 植物学报, 2022, 57(1): 12-23. |
| [6] | 周俭民. 免疫信号轴揭示水稻与病原菌斗争的秘密[J]. 植物学报, 2021, 56(5): 513-515. |
| [7] | 王伟, 唐定中. 两类免疫受体强强联手筑牢植物免疫防线[J]. 植物学报, 2021, 56(2): 142-146. |
| [8] | 杨程惠子,唐先宇,李威,夏石头. NLR及其在植物抗病中的调控作用[J]. 植物学报, 2020, 55(4): 497-504. |
| [9] | 崔亚宁, 钱虹萍, 赵艳霞, 李晓娟. 模式识别受体的胞内转运及其在植物免疫中的作用[J]. 植物学报, 2020, 55(3): 329-339. |
| [10] | 李伟滔, 贺闽, 陈学伟. ZmFBL41 Chang7-2: 玉米抗纹枯病的关键利器[J]. 植物学报, 2019, 54(5): 547-549. |
| [11] | 夏石头, 李昕. 开启防御之门: 植物抗病小体[J]. 植物学报, 2019, 54(3): 288-292. |
| [12] | 闫佳, 刘雅琼, 侯岁稳. 植物抗病蛋白研究进展[J]. 植物学报, 2018, 53(2): 250-263. |
| [13] | 牛国清 邓宇. 天蚕素及其在植物抗病育种中的应用[J]. 植物学报, 2003, 20(04): 476-480. |
| [14] | 王红 叶和春 李国凤. 诱导子的作用方式及其在植物组织培养中的应用[J]. 植物学报, 1999, 16(01): 11-18. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||