

植物学报 ›› 2020, Vol. 55 ›› Issue (2): 147-162.DOI: 10.11983/CBB19103 cstr: 32102.14.CBB19103
收稿日期:2019-05-30
接受日期:2019-09-19
出版日期:2020-03-01
发布日期:2020-02-12
通讯作者:
许杰
基金资助:
Zeyuan Zuo,Wanlin Liu,Jie Xu( )
)
Received:2019-05-30
Accepted:2019-09-19
Online:2020-03-01
Published:2020-02-12
Contact:
Jie Xu
摘要: 在植物基因组中, 除了同源基因成簇现象外, 近年来还发现一些具有共表达特性的异源基因也能够以基因簇形式存在, 但这些异源基因簇的进化和生物学功能尚不清楚。花药发育和花粉形成是植物进化出的特有的生殖生物学过程, 同时产生了一些在花药绒毡层中特异表达和特定功能的基因簇基因。该研究通过筛选和分析花药绒毡层中基因簇基因的分子特性、表达调控、基因年龄和基因重复进化等信息, 探讨花药基因簇基因与植物开花功能进化之间的关系。结果表明, 在拟南芥(Arabidopsis thaliana)中共筛选到84个(13个基因簇)花药绒毡层特异高表达的基因簇基因, 它们主要产生于串联重复事件, 76%的基因出现在开花植物分化后的阶段, 主要参与生殖发育、花粉鞘组成和脂代谢等生物学过程。研究初步解析了拟南芥花药绒毡层中基因簇基因的基本特征、生物学功能和基因进化机制, 为深入揭示植物基因簇基因的遗传学功能奠定了基础。
左泽远,刘琬琳,许杰. 拟南芥花药绒毡层细胞中具有基因簇特征的基因进化和功能分析. 植物学报, 2020, 55(2): 147-162.
Zeyuan Zuo,Wanlin Liu,Jie Xu. Evolution and Functional Analysis of Gene Clusters in Anther Tapetum Cells of Arabidopsis thaliana. Chinese Bulletin of Botany, 2020, 55(2): 147-162.
| Cluster No. | AGI | MW (kDa) | PI | Location | TM | Description |
|---|---|---|---|---|---|---|
| 1 | AT1G12060 | 24786.8 | 9.09 | Undefined | 0 | BCL-2-associated athanogene 5 |
| 1 | AT1G12064 | 11961.3 | 9.24 | Secreted | 1 | Unknown protein |
| 1 | AT1G12070 | 25456.4 | 4.52 | Other | 0 | Immunoglobulin E-set superfamily protein |
| 1 | AT1G12080 | 15381.7 | 3.74 | Undefined | 0 | Vacuolar calcium-binding protein-related |
| 1 | AT1G12090 | 13912.3 | 8.15 | Secreted | 0 | Extensin-like protein |
| 1 | AT1G12100 | 13909.2 | 6.90 | Secreted | 0 | Bifunctional inhibitor/lipid-transfer protein/seed storage |
| 1 | AT1G12110 | 64921.2 | 8.57 | Other | 11 | Nitrate transporter 1.1 |
| 2 | AT1G61560 | 67225.5 | 10.06 | Undefined | 8 | Seven transmembrane MLO family protein |
| 2 | AT1G61563 | 8906.4 | 9.93 | Secreted | 1 | Ralf-like 8 |
| 2 | AT1G61566 | 8259.6 | 9.93 | Secreted | 1 | Ralf-like 9 |
| 2 | AT1G61570 | 9411.6 | 5.03 | Undefined | 0 | Translocase of the inner mitochondrial membrane 13 |
| 2 | AT1G61575 | 5641.3 | 4.50 | Undefined | 0 | Unknown protein |
| 3 | AT1G74540 | 56740.6 | 8.36 | Undefined | 1 | Cytochrome P450, family 98 |
| 3 | AT1G74550 | 55418.9 | 8.88 | Undefined | 0 | Cytochrome P450, family 98 |
| 4 | AT1G75880 | 41333.3 | 8.23 | Secreted | 1 | SGNH hydrolase-type esterase |
| 4 | AT1G75890 | 42126.2 | 8.44 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75900 | 39815.4 | 5.47 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75910 | 37910.6 | 10.30 | Secreted | 0 | Extracellular lipase 4 |
| 4 | AT1G75920 | 39254.5 | 10.06 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75930 | 38595.7 | 9.89 | Secreted | 0 | Extracellular lipase 6 |
| 4 | AT1G75940 | 61674.7 | 6.04 | Secreted | 1 | Glycosyl hydrolase superfamily protein |
| 5 | AT2G47010 | 55049.8 | 6.50 | Secreted | 0 | Unknown protein |
| 5 | AT2G47020 | 46827.8 | 7.36 | Mitochondrion | 0 | Peptide chain release factor 1 |
| 5 | AT2G47030 | 64137.1 | 9.44 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47040 | 64727.7 | 9.32 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47050 | 23775.5 | 9.35 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47060 | 43753.3 | 7.67 | Other | 0 | Protein kinase superfamily protein |
| 6 | AT3G07810 | 52055.5 | 8.81 | Other | 0 | RNA-binding (RRM/RBD/RNP motifs) family protein |
| 6 | AT3G07820 | 41681.1 | 7.34 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 6 | AT3G07830 | 42912.9 | 8.69 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 6 | AT3G07840 | 42591.5 | 8.08 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 6 | AT3G07850 | 45599.9 | 8.43 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 6 | AT3G07860 | 18672.5 | 10.45 | Other | 0 | Ubiquitin-like superfamily protein |
| 7 | AT3G28730 | 71645.6 | 5.56 | Undefined | 0 | High mobility group |
| 7 | AT3G28740 | 57691.8 | 9.23 | Secreted | 1 | Cytochrome P450 superfamily protein |
| 7 | AT3G28750 | 35478.4 | 10.45 | Secreted | 0 | Unknown protein |
| 7 | AT3G28760 | 49598.7 | 6.72 | Undefined | 0 | Unknown protein |
| 7 | AT3G28770 | 232851.8 | 4.73 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28780 | 61329 | 3.92 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28790 | 62765.8 | 10.04 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28810 | 47872.2 | 9.70 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28820 | 47786 | 9.38 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28830 | 55500.6 | 10.42 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28840 | 36875.3 | 10.06 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28850 | 48178.4 | 4.72 | Undefined | 0 | Glutaredoxin family protein |
| 7 | AT3G28860 | 136786.9 | 8.37 | Other | 10 | ATP binding cassette subfamily B19 |
| 8 | AT4G29290 | 8554.9 | 6.92 | Secreted | 1 | Low-molecular-weight cysteine-rich 26 |
| 8 | AT4G29300 | 8510.8 | 4.40 | Secreted | 1 | Low-molecular-weight cysteine-rich 27 |
| 8 | AT4G29305 | 8359.7 | 7.91 | Secreted | 1 | Low-molecular-weight cysteine-rich 25 |
| 8 | AT4G29310 | 45655.6 | 9.63 | Undefined | 0 | Protein of unknown function (DUF1005) |
| 8 | AT4G29330 | 29213.2 | 10.33 | Other | 6 | DERLIN-1 |
| 8 | AT4G29340 | 14418.3 | 4.84 | Other | 0 | Profilin 4 |
| 8 | AT4G29350 | 13997.8 | 4.67 | Other | 0 | Profilin 2 |
| 9 | AT5G07410 | 39336.9 | 8.70 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 9 | AT5G07420 | 39658 | 9.40 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 9 | AT5G07430 | 39910.3 | 9.11 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 9 | AT5G07440 | 44698.7 | 6.51 | Undefined | 0 | Glutamate dehydrogenase 2 |
| 9 | AT5G07450 | 25004.5 | 6.01 | Undefined | 0 | Cyclin p4;3 |
| 10 | AT5G07510 | 18462.4 | 12.43 | Secreted | 3 | Glycine-rich protein 14 |
| 10 | AT5G07520 | 21482.5 | 10.55 | Secreted | 3 | Glycine-rich protein 18 |
| 10 | AT5G07530 | 53192.9 | 11.13 | Undefined | 3 | Glycine rich protein 17 |
| 10 | AT5G07540 | 22300.1 | 10.85 | Secreted | 3 | Glycine-rich protein 16 |
| 10 | AT5G07550 | 10682.5 | 10.13 | Secreted | 2 | Glycine-rich protein 19 |
| 10 | AT5G07560 | 15458.3 | 11.20 | Other | 3 | Glycine-rich protein 20 |
| 10 | AT5G07570 | 148459.7 | 3.36 | Secreted | 10 | Glycine/proline-rich protein |
| 11 | AT5G17450 | 16995.5 | 10.18 | Undefined | 0 | Heavy metal transport/detoxification |
| 11 | AT5G17460 | 35918.8 | 8.34 | Undefined | 0 | Unknown protein |
| 11 | AT5G17470 | 16320.1 | 4.01 | Other | 0 | EF hand calcium-binding protein family |
| 11 | AT5G17480 | 9048 | 4.30 | Other | 0 | Pollen calcium-binding protein 1 |
| 11 | AT5G17490 | 57326.1 | 4.49 | Other | 0 | RGA-like protein 3 |
| 11 | AT5G17500 | 59551 | 8.19 | Secreted | 1 | Glycosyl hydrolase superfamily protein |
| 12 | AT5G53800 | 41786.3 | 10.38 | Other | 0 | Unknown protein |
| 12 | AT5G53810 | 41944.2 | 5.17 | Other | 0 | O-methyltransferase family protein |
| 12 | AT5G53820 | 7028.7 | 10.26 | Undefined | 0 | Late embryogenesis abundant protein (LEA) family protein |
| 12 | AT5G53830 | 26918.8 | 10.18 | Undefined | 0 | VQ motif-containing protein |
| 12 | AT5G53840 | 50906.6 | 7.31 | Other | 0 | F-box/RNI-like/FBD-like domains-containing pro- tein |
| 12 | AT5G53850 | 56520.1 | 6.14 | Undefined | 0 | Haloacid dehalogenase-like hydrolase family protein |
| 13 | AT5G67460 | 41995.4 | 8.16 | Secreted | 0 | O-glycosyl hydrolases family 17 protein |
| 13 | AT5G67470 | 98648.8 | 9.91 | Secreted | 2 | Formin homolog 6 |
| 13 | AT5G67480 | 43817.7 | 9.59 | Undefined | 0 | BTB and TAZ domain protein 4 |
| 13 | AT5G67490 | 11869.8 | 4.25 | Undefined | 0 | Unknown protein |
| 13 | AT5G67500 | 32904.9 | 7.68 | Other | 0 | Voltage dependent anion channel 2 |
| 13 | AT5G67510 | 16790.3 | 11.74 | Undefined | 0 | Translation protein SH3-like family protein |
| 13 | AT5G67520 | 34064.3 | 9.29 | Undefined | 0 | Adenosine-5'-phosphosulfate (APS) kinase 4 |
表1 拟南芥花药发育中特异性表达基因簇基因的基本信息
Table 1 Basic information of anther-specific expressed cluster genes during Arabidopsis anther development
| Cluster No. | AGI | MW (kDa) | PI | Location | TM | Description |
|---|---|---|---|---|---|---|
| 1 | AT1G12060 | 24786.8 | 9.09 | Undefined | 0 | BCL-2-associated athanogene 5 |
| 1 | AT1G12064 | 11961.3 | 9.24 | Secreted | 1 | Unknown protein |
| 1 | AT1G12070 | 25456.4 | 4.52 | Other | 0 | Immunoglobulin E-set superfamily protein |
| 1 | AT1G12080 | 15381.7 | 3.74 | Undefined | 0 | Vacuolar calcium-binding protein-related |
| 1 | AT1G12090 | 13912.3 | 8.15 | Secreted | 0 | Extensin-like protein |
| 1 | AT1G12100 | 13909.2 | 6.90 | Secreted | 0 | Bifunctional inhibitor/lipid-transfer protein/seed storage |
| 1 | AT1G12110 | 64921.2 | 8.57 | Other | 11 | Nitrate transporter 1.1 |
| 2 | AT1G61560 | 67225.5 | 10.06 | Undefined | 8 | Seven transmembrane MLO family protein |
| 2 | AT1G61563 | 8906.4 | 9.93 | Secreted | 1 | Ralf-like 8 |
| 2 | AT1G61566 | 8259.6 | 9.93 | Secreted | 1 | Ralf-like 9 |
| 2 | AT1G61570 | 9411.6 | 5.03 | Undefined | 0 | Translocase of the inner mitochondrial membrane 13 |
| 2 | AT1G61575 | 5641.3 | 4.50 | Undefined | 0 | Unknown protein |
| 3 | AT1G74540 | 56740.6 | 8.36 | Undefined | 1 | Cytochrome P450, family 98 |
| 3 | AT1G74550 | 55418.9 | 8.88 | Undefined | 0 | Cytochrome P450, family 98 |
| 4 | AT1G75880 | 41333.3 | 8.23 | Secreted | 1 | SGNH hydrolase-type esterase |
| 4 | AT1G75890 | 42126.2 | 8.44 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75900 | 39815.4 | 5.47 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75910 | 37910.6 | 10.30 | Secreted | 0 | Extracellular lipase 4 |
| 4 | AT1G75920 | 39254.5 | 10.06 | Secreted | 0 | GDSL-like lipase/Acylhydrolase |
| 4 | AT1G75930 | 38595.7 | 9.89 | Secreted | 0 | Extracellular lipase 6 |
| 4 | AT1G75940 | 61674.7 | 6.04 | Secreted | 1 | Glycosyl hydrolase superfamily protein |
| 5 | AT2G47010 | 55049.8 | 6.50 | Secreted | 0 | Unknown protein |
| 5 | AT2G47020 | 46827.8 | 7.36 | Mitochondrion | 0 | Peptide chain release factor 1 |
| 5 | AT2G47030 | 64137.1 | 9.44 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47040 | 64727.7 | 9.32 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47050 | 23775.5 | 9.35 | Secreted | 1 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| 5 | AT2G47060 | 43753.3 | 7.67 | Other | 0 | Protein kinase superfamily protein |
| 6 | AT3G07810 | 52055.5 | 8.81 | Other | 0 | RNA-binding (RRM/RBD/RNP motifs) family protein |
| 6 | AT3G07820 | 41681.1 | 7.34 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 6 | AT3G07830 | 42912.9 | 8.69 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 6 | AT3G07840 | 42591.5 | 8.08 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 6 | AT3G07850 | 45599.9 | 8.43 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 6 | AT3G07860 | 18672.5 | 10.45 | Other | 0 | Ubiquitin-like superfamily protein |
| 7 | AT3G28730 | 71645.6 | 5.56 | Undefined | 0 | High mobility group |
| 7 | AT3G28740 | 57691.8 | 9.23 | Secreted | 1 | Cytochrome P450 superfamily protein |
| 7 | AT3G28750 | 35478.4 | 10.45 | Secreted | 0 | Unknown protein |
| 7 | AT3G28760 | 49598.7 | 6.72 | Undefined | 0 | Unknown protein |
| 7 | AT3G28770 | 232851.8 | 4.73 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28780 | 61329 | 3.92 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28790 | 62765.8 | 10.04 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28810 | 47872.2 | 9.70 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28820 | 47786 | 9.38 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28830 | 55500.6 | 10.42 | Secreted | 1 | Protein of unknown function (DUF1216) |
| 7 | AT3G28840 | 36875.3 | 10.06 | Secreted | 0 | Protein of unknown function (DUF1216) |
| 7 | AT3G28850 | 48178.4 | 4.72 | Undefined | 0 | Glutaredoxin family protein |
| 7 | AT3G28860 | 136786.9 | 8.37 | Other | 10 | ATP binding cassette subfamily B19 |
| 8 | AT4G29290 | 8554.9 | 6.92 | Secreted | 1 | Low-molecular-weight cysteine-rich 26 |
| 8 | AT4G29300 | 8510.8 | 4.40 | Secreted | 1 | Low-molecular-weight cysteine-rich 27 |
| 8 | AT4G29305 | 8359.7 | 7.91 | Secreted | 1 | Low-molecular-weight cysteine-rich 25 |
| 8 | AT4G29310 | 45655.6 | 9.63 | Undefined | 0 | Protein of unknown function (DUF1005) |
| 8 | AT4G29330 | 29213.2 | 10.33 | Other | 6 | DERLIN-1 |
| 8 | AT4G29340 | 14418.3 | 4.84 | Other | 0 | Profilin 4 |
| 8 | AT4G29350 | 13997.8 | 4.67 | Other | 0 | Profilin 2 |
| 9 | AT5G07410 | 39336.9 | 8.70 | Secreted | 1 | Pectin lyase-like superfamily protein |
| 9 | AT5G07420 | 39658 | 9.40 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 9 | AT5G07430 | 39910.3 | 9.11 | Secreted | 0 | Pectin lyase-like superfamily protein |
| 9 | AT5G07440 | 44698.7 | 6.51 | Undefined | 0 | Glutamate dehydrogenase 2 |
| 9 | AT5G07450 | 25004.5 | 6.01 | Undefined | 0 | Cyclin p4;3 |
| 10 | AT5G07510 | 18462.4 | 12.43 | Secreted | 3 | Glycine-rich protein 14 |
| 10 | AT5G07520 | 21482.5 | 10.55 | Secreted | 3 | Glycine-rich protein 18 |
| 10 | AT5G07530 | 53192.9 | 11.13 | Undefined | 3 | Glycine rich protein 17 |
| 10 | AT5G07540 | 22300.1 | 10.85 | Secreted | 3 | Glycine-rich protein 16 |
| 10 | AT5G07550 | 10682.5 | 10.13 | Secreted | 2 | Glycine-rich protein 19 |
| 10 | AT5G07560 | 15458.3 | 11.20 | Other | 3 | Glycine-rich protein 20 |
| 10 | AT5G07570 | 148459.7 | 3.36 | Secreted | 10 | Glycine/proline-rich protein |
| 11 | AT5G17450 | 16995.5 | 10.18 | Undefined | 0 | Heavy metal transport/detoxification |
| 11 | AT5G17460 | 35918.8 | 8.34 | Undefined | 0 | Unknown protein |
| 11 | AT5G17470 | 16320.1 | 4.01 | Other | 0 | EF hand calcium-binding protein family |
| 11 | AT5G17480 | 9048 | 4.30 | Other | 0 | Pollen calcium-binding protein 1 |
| 11 | AT5G17490 | 57326.1 | 4.49 | Other | 0 | RGA-like protein 3 |
| 11 | AT5G17500 | 59551 | 8.19 | Secreted | 1 | Glycosyl hydrolase superfamily protein |
| 12 | AT5G53800 | 41786.3 | 10.38 | Other | 0 | Unknown protein |
| 12 | AT5G53810 | 41944.2 | 5.17 | Other | 0 | O-methyltransferase family protein |
| 12 | AT5G53820 | 7028.7 | 10.26 | Undefined | 0 | Late embryogenesis abundant protein (LEA) family protein |
| 12 | AT5G53830 | 26918.8 | 10.18 | Undefined | 0 | VQ motif-containing protein |
| 12 | AT5G53840 | 50906.6 | 7.31 | Other | 0 | F-box/RNI-like/FBD-like domains-containing pro- tein |
| 12 | AT5G53850 | 56520.1 | 6.14 | Undefined | 0 | Haloacid dehalogenase-like hydrolase family protein |
| 13 | AT5G67460 | 41995.4 | 8.16 | Secreted | 0 | O-glycosyl hydrolases family 17 protein |
| 13 | AT5G67470 | 98648.8 | 9.91 | Secreted | 2 | Formin homolog 6 |
| 13 | AT5G67480 | 43817.7 | 9.59 | Undefined | 0 | BTB and TAZ domain protein 4 |
| 13 | AT5G67490 | 11869.8 | 4.25 | Undefined | 0 | Unknown protein |
| 13 | AT5G67500 | 32904.9 | 7.68 | Other | 0 | Voltage dependent anion channel 2 |
| 13 | AT5G67510 | 16790.3 | 11.74 | Undefined | 0 | Translation protein SH3-like family protein |
| 13 | AT5G67520 | 34064.3 | 9.29 | Undefined | 0 | Adenosine-5'-phosphosulfate (APS) kinase 4 |
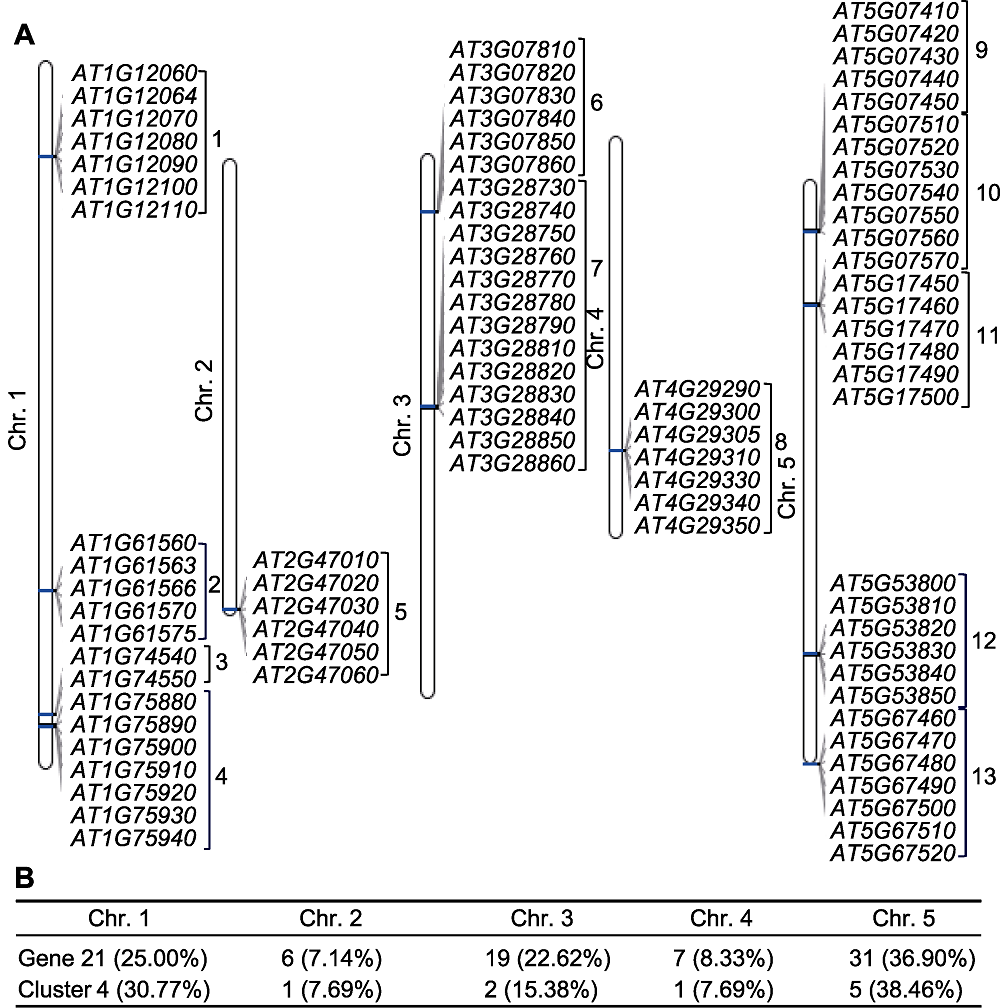
图2 拟南芥花药发育特异表达的84个候选基因的染色体分布 (A) 候选基因在1-5号染色体上的分布情况(数字代表基因所属的基因簇); (B) 候选基因在染色体上具体分布的数目及比例
Figure 2 Chromosome distribution of anther-specific expressed 84 candidate genes during Arabidopsis anther development (A) Distribution of candidate genes on 1-5 chromosome (Numbers represent which cluster genes belong to); (B) Number and proportion of candidate genes distributed on chromosome
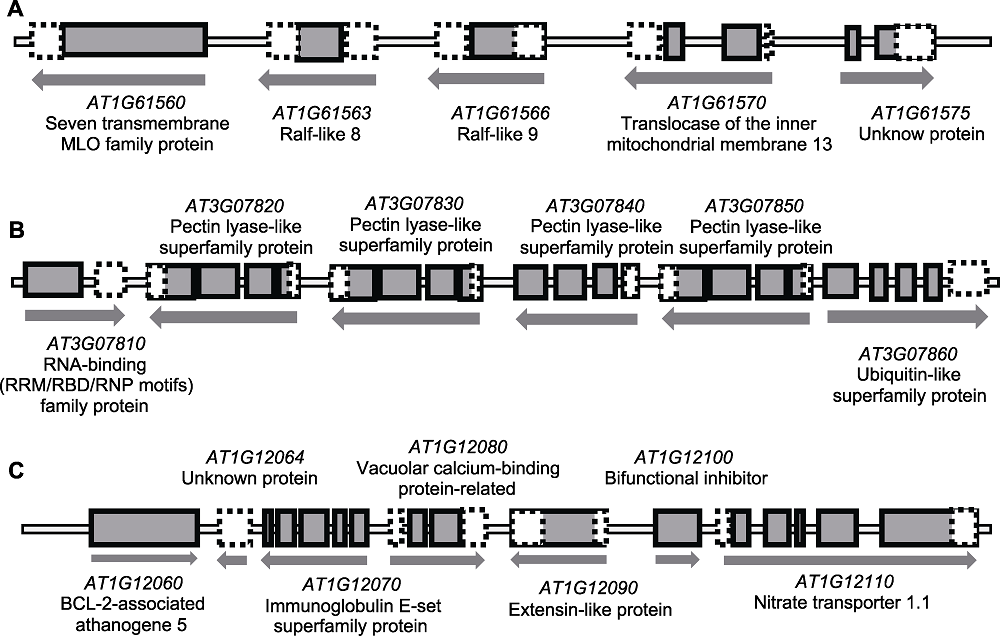
图3 拟南芥花药发育特异表达基因簇基因的结构和转录方向 (A) Ralf-like家族基因簇基因结构; (B) Pectin lyase-like家族基因簇基因结构; (C) AT1G12060-AT1G12110基因簇基因结构。图中包含非编码区域和未知蛋白(虚线白框)、外显子(黑边灰框)、内含子(外显子之间的间隔)、转录方向(下方黑色箭头)和基因名注释。长度按照实际长度等比例近似缩放。
Figure 3 Gene structure and transcriptional direction of anther-specific expressed clustered genes during Arabidopsis anther development (A) Gene structure of Ralf-like family gene cluster; (B) Gene structure of pectin lyase-like family gene cluster; (C) Gene structure of AT1G12060-AT1G12110 gene cluster. Dotted line with white square: untranslated region and unknown protein; Black side with grey square: exon; Interval between exons: intron; Black arrow below: transcription direction and gene annotations. Size of gene is approximately proportional to the actual size.
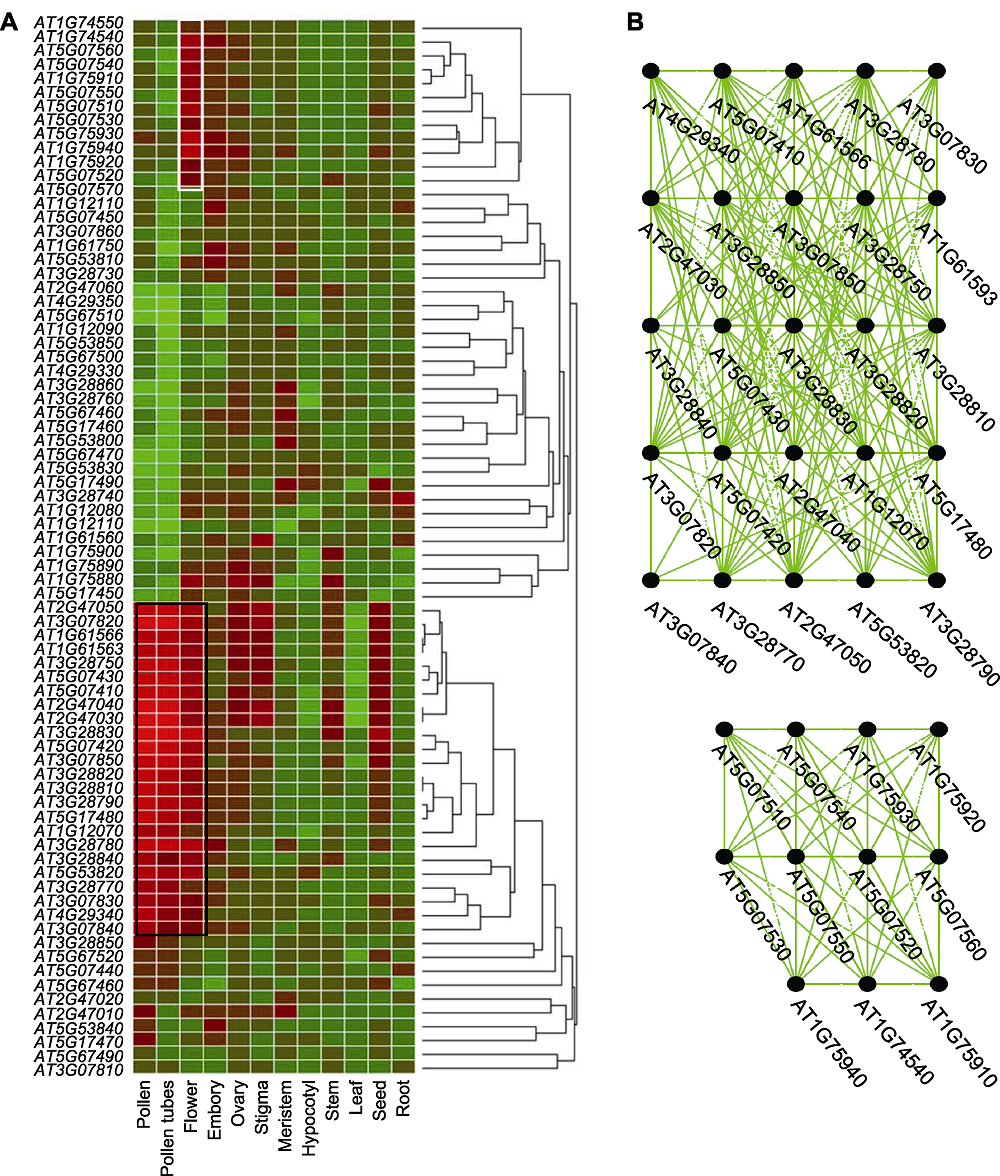
图4 拟南芥花药发育特异表达候选基因的表达分析 (A) 表达聚类热图, 组织部位包括根、种子、叶、茎、子叶下轴、分生组织、柱头、子房、胚、花、花粉管和花粉; (B) 共表达网络
Figure 4 Expression-pattern analysis of anther-specific expressed candidate genes during Arabidopsis anther development (A) Clustered expression heatmap, containing tissues such as root, seed, leaf, stem, hypocotyl, meristem, stigma, ovary, embryo, flower, pollen tube, and pollen; (B) Co-expression network
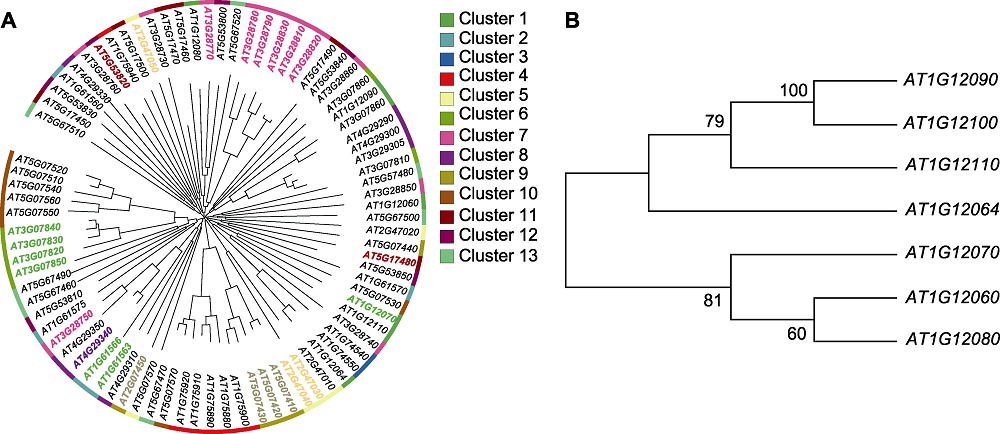
图5 拟南芥花药发育特异表达候选基因进化树 (A) 候选基因进化树上彩色的基因ID代表初步筛选的24个特异性高表达基因, 外侧的颜色条代表筛选的84个基因, 同一颜色带属于同一基因簇; (B) 1号基因簇进化树, 分支上的数字来自bootstrap验证。
Figure 5 Phylogenetic trees of anther-specific expressed candidate genes during Arabidopsis anther development (A) Phylogenetic tree of candidate genes, the colored gene ID in phylogenetic tree represents 24 primarily screened specific high-expression genes, outside color strips represent 13 clusters, with 84 screened genes, and genes under the same color belonging to same cluster; (B) Phylogenetic tree of gene cluster 1, numbers on branches represent bootstrap validation.
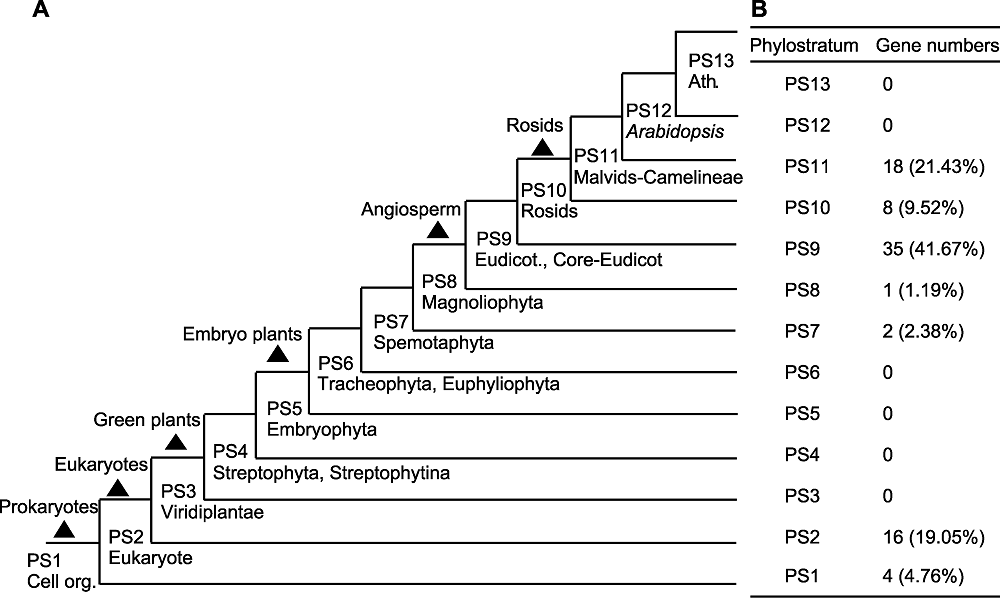
图6 拟南芥系统发生断层学分析结果 (A) 拟南芥基因组进化层图, 包括原核生物、真核生物、绿色植物、胚胎植物、被子植物和蔷薇植物; (B) 候选基因在PS1-PS13层级的分布及比例
Figure 6 Phylostratigraphic profiles of Arabidopsis genes (A) Phylostratigraphic map of Arabidopsis, include prokaryotes, eukaryotes, green plants, embryo plants, angiosperm and rosids; (B) Distribution and proportion of candidate genes in Phylostratum 1-13
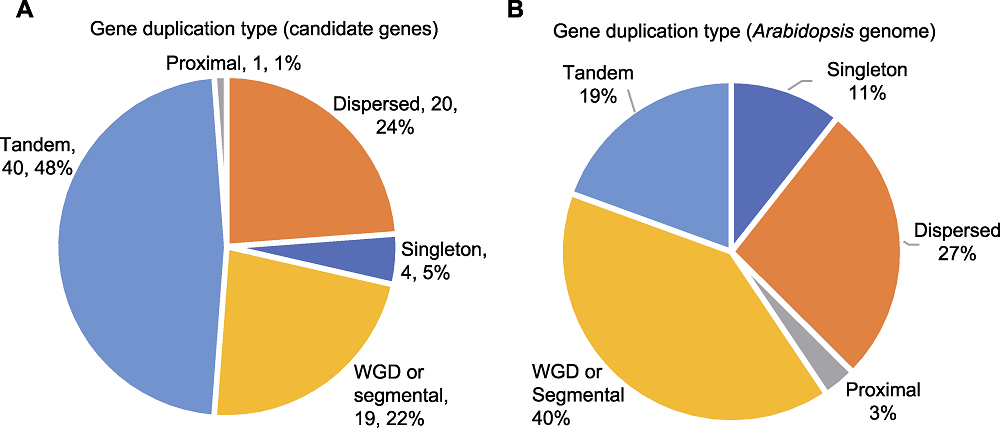
图7 拟南芥基因重复事件 (A) 候选基因重复事件; (B) 拟南芥基因组中基因重复事件。Proximal: 邻域重复事件; Dispersed: 分散重复事件; Singleton: 非重复事件; Tandem: 串联重复事件; WGD or segmental: 全基因组或片段重复事件
Figure 7 Gene duplication events in Arabidopsis (A) Gene duplication events of candidate genes; (B) Gene duplication events in Arabidopsis genome. Proximal: Proximal duplication event; Dispersed: Dispersed duplication event; Singleton: No duplication event; Tandem: Tandem duplication event; WGD or segmental: Whole genome duplication or segmental duplication event
| Phylostratum | WGD | Tandem | Proximal | Dispersed | Singleton |
|---|---|---|---|---|---|
| PS13 | 0 | 0 | 0 | 0 | 0 |
| PS12 | 0 | 0 | 0 | 0 | 0 |
| PS11 | 4 (21.05%) | 13 (32.50%) | 0 | 1 (5.00%) | 0 |
| PS10 | 4 (21.05%) | 2 (5.00%) | 0 | 1 (5.00%) | 1 (25.00%) |
| PS9 | 7 (36.84%) | 15 (37.50%) | 0 | 11 (55.00%) | 2 (50.00%) |
| PS8 | 0 | 1 (2.50%) | 0 | 0 | 0 |
| PS7 | 1 (5.26%) | 0 | 0 | 1 (5.00%) | 0 |
| PS6 | 0 | 0 | 0 | 0 | 0 |
| PS5 | 0 | 0 | 0 | 0 | 0 |
| PS4 | 0 | 0 | 0 | 0 | 0 |
| PS3 | 0 | 0 | 0 | 0 | 0 |
| PS2 | 3 (15.79%) | 7 (17.50%) | 0 | 6 (30.00%) | 0 |
| PS1 | 0 | 2 (5.00%) | 1 (100%) | 0 | 1 (25.00%) |
表2 拟南芥基因进化层与基因重复事件的关系
Table 2 Relationships between phylostratum and gene duplication events in Arabidopsis
| Phylostratum | WGD | Tandem | Proximal | Dispersed | Singleton |
|---|---|---|---|---|---|
| PS13 | 0 | 0 | 0 | 0 | 0 |
| PS12 | 0 | 0 | 0 | 0 | 0 |
| PS11 | 4 (21.05%) | 13 (32.50%) | 0 | 1 (5.00%) | 0 |
| PS10 | 4 (21.05%) | 2 (5.00%) | 0 | 1 (5.00%) | 1 (25.00%) |
| PS9 | 7 (36.84%) | 15 (37.50%) | 0 | 11 (55.00%) | 2 (50.00%) |
| PS8 | 0 | 1 (2.50%) | 0 | 0 | 0 |
| PS7 | 1 (5.26%) | 0 | 0 | 1 (5.00%) | 0 |
| PS6 | 0 | 0 | 0 | 0 | 0 |
| PS5 | 0 | 0 | 0 | 0 | 0 |
| PS4 | 0 | 0 | 0 | 0 | 0 |
| PS3 | 0 | 0 | 0 | 0 | 0 |
| PS2 | 3 (15.79%) | 7 (17.50%) | 0 | 6 (30.00%) | 0 |
| PS1 | 0 | 2 (5.00%) | 1 (100%) | 0 | 1 (25.00%) |
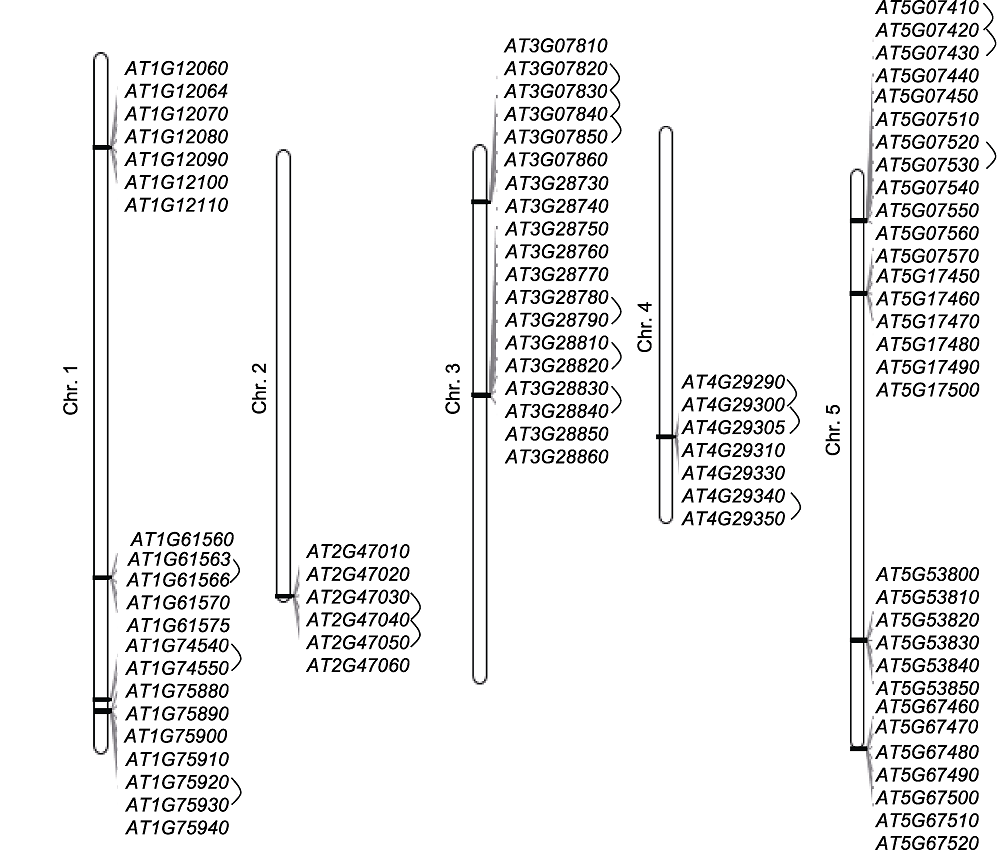
图8 拟南芥花药发育特异表达候选基因基因簇分布及内部串联重复关系 Chr.1-5代表1-5号染色体。黑色连线代表存在串联重复关系。
Figure 8 Distribution of gene clusters and tandem duplication relationships of anther-specific expressed candidate genes in Arabidopsis anther development Chr.1-5 represents 1-5 chromosome. Black links represent they have tandem duplication relationships.
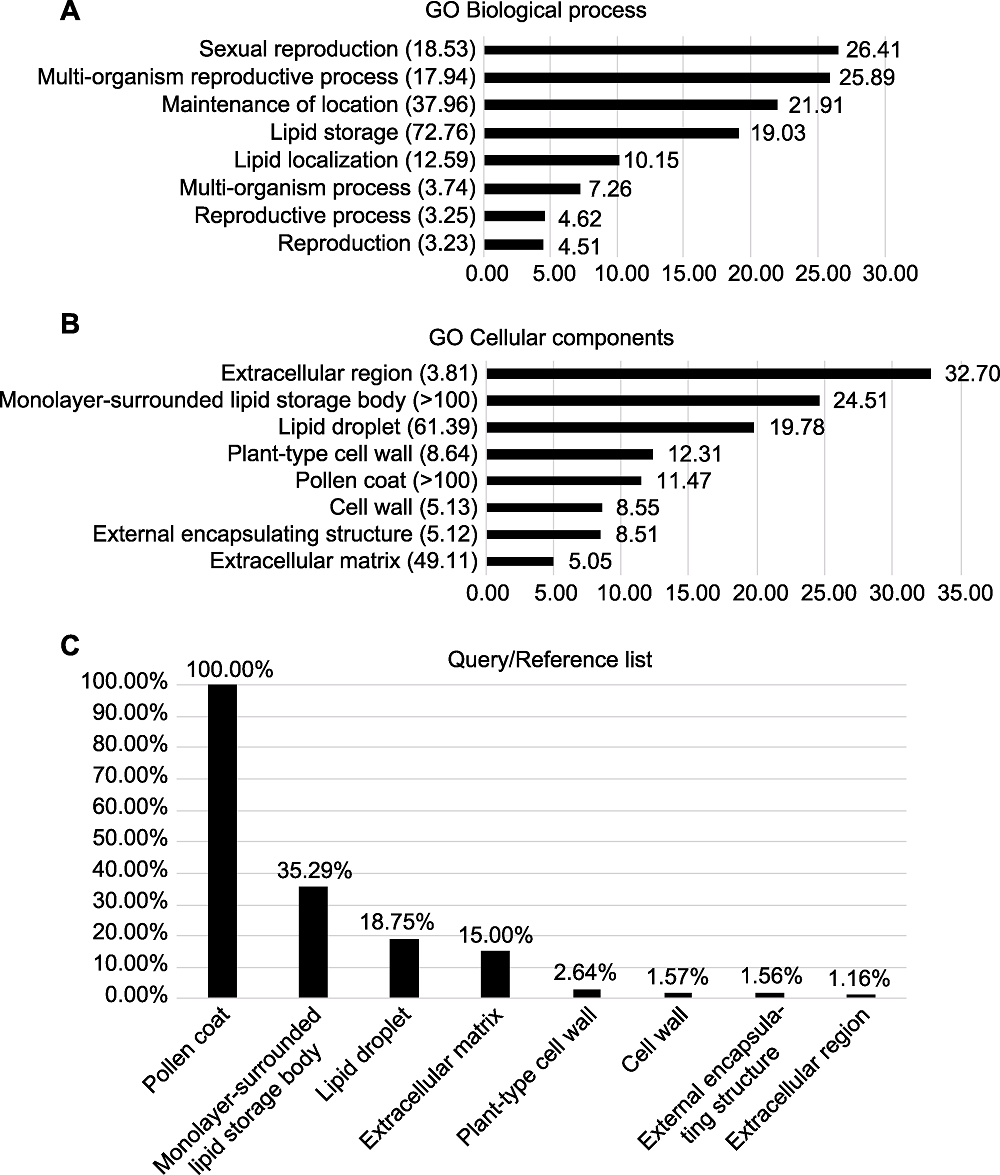
图9 拟南芥花药发育特异表达候选基因GO (gene ontology)富集分析 (A) GO生物过程富集分析; (B) GO细胞组分富集分析(图(A)和(B)纵坐标为GO项目注释, 括号中是富集倍数, 横坐标为P值负对数); (C) GO细胞组成后续分析(纵坐标为候选基因中存在GO项目的基因数目和拟南芥全基因组中存在GO项目的基因数目比值, 横坐标为GO项目的注释)
Figure 9 Gene ontology (GO) enrichment analysis of anther-specific expressed candidate genes during Arabidopsis anther development (A) GO biological process enrichment analysis; (B) GO cellular component enrichment analysis (Figures (A) and (B) y-axis represents annotations of GO terms, number in parenthesis represents GO enrichment fold and x-axis represents minus logarithm of P value); (C) Subsequent analysis of GO cellular components (y-axis represents number of genes in a GO term in candidate genes to number of genes in a GO term in Arabidopsis genome and x-axis represents annotation of GO terms)
| [1] | 李鸿健, 谭军 ( 2006). 基因倍增研究进展. 生命科学 18, 150-154. |
| [2] | 吕海舟, 刘琬菁, 何柳, 徐志超, 罗红梅 ( 2017). 植物次生代谢基因簇研究进展. 植物科学学报 35, 609-621. |
| [3] | 孙红正, 葛颂 ( 2010). 重复基因的进化——回顾与进展. 植物学报 45, 13-22. |
| [4] | 朱子超, 柴友荣, 杨光伟 ( 2009). 查尔酮合酶基因重复-歧化的遗传学效应. 安徽农学通报 15, 25-30. |
| [5] | Carbon S, Ireland A, Mungall CJ, Shu SQ, Marshall B, Lewis S , the AmiGO Hub, the Web Presence Working Group ( 2009). AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288-289. |
| [6] | Chen CJ, Xia R, Chen H, He YH ( 2018). TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. doi: 10.1101/289660. |
| [7] | Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA ( 2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412-421. |
| [8] | Cui X, Lv Y, Chen ML, Nikoloski Z, Twell D, Zhang DB ( 2015). Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol Plant 8, 935-945. |
| [9] | De Bodt S, Maere S, Van de Peer Y ( 2005). Genome duplication and the origin of angiosperms. Trends Ecol Evol 20, 591-597. |
| [10] | Díez B, Gutiérrez S, Barredo JL, van Solingen P, van der Voort LH, Martín JF ( 1990). The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl- valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem 265, 16358-16365. |
| [11] | Domazet-Lošo T, Brajković J, Tautz D ( 2007). A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23, 533-539. |
| [12] | Domazet-Lošo T, Tautz D ( 2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468, 815-818. |
| [13] | Falara V, Akhtar TA, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini ME, Schilmiller AL, Last RL, Schuurink RC, Pichersky E ( 2011). The tomato terpene synthase gene family. Plant Physiol 157, 770-789. |
| [14] | Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J ( 1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531-1545. |
| [15] | Guenzi E, Galli G, Grgurina I, Gross DC, Grandi G ( 1998). Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J Biol Chem 273, 32857-32863. |
| [16] | Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, Tikunov Y, Bovy A, Chikate Y, Singh P, Rogachev I, Beekwilder J, Giri AP, Aharoni A ( 2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175-179. |
| [17] | Klein J, Figueroa F ( 1986). Evolution of the major histocompatibility complex. Crit Rev Immunol 6, 295-386. |
| [18] | Koch MA, Haubold B, Mitchell-Olds T ( 2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17, 1483-1498. |
| [19] | Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV ( 2002). Selection in the evolution of gene duplications. Genome Biol 3, research0008. |
| [20] | Kumar S, Stecher G, Tamura K ( 2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870-1874. |
| [21] | Li DD, Xue JS, Zhu J, Yang ZN ( 2017). Gene regulatory network for tapetum development in Arabidopsis thaliana. Front Plant Sci 8, 1559. |
| [22] | Lora J, Testillano PS, Risueño MC, Hormaza JI, Herrero M ( 2009). Pollen development in Annona cherimola Mill.(Annonaceae). Implications for the evolution of aggregated pollen. BMC Plant Biol 9, 129. |
| [23] | MacCabe AP, Riach MB, Unkles SE, Kinghorn JR ( 1990). The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J 9, 279-287. |
| [24] | Mascarenhas JP ( 1990). Gene activity during pollen development. Annu Rev Plant Physiol Plant Mol Biol 41, 317-338. |
| [25] | Mi HY, Huang XS, Muruganujan A, Tang HM, Mills C, Kang D, Thomas PD ( 2017). PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183-D189. |
| [26] | Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H ( 1999). A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res 9, 1175-1183. |
| [27] | Prince VE, Pickett FB ( 2002). Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3, 827-837. |
| [28] | Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A ( 2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101, 8233-8238. |
| [29] | Slot JC ( 2017). Fungal gene cluster diversity and evolution. Adv Genet 100, 141-178. |
| [30] | Thornton J ( 2006). New genes, new functions: gene family evolution and phylogenetics. In: Fox CW, Wolf JB, eds. Evolutionary Genetics: Concepts and Case Studies. New York: Oxford University Press. pp. 157-172. |
| [31] | Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, Kissinger JC, Paterson AH ( 2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40, e49. |
| [32] | Wilson ZA, Zhang DB ( 2009). From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60, 1479-1492. |
| [33] | Worthen DB ( 2008). Streptomyces in nature and medicine: the antibiotic makers. J History Med Allied Sci 63, 273-274. |
| [34] | Yang SL, Xie LF, Mao HZ, San Puah C, Yang WC, Jiang LX, Sundaresan V, Ye D ( 2003). TAPETUM DETERMINANT 1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15, 2792-2804. |
| [35] | Zhang JZ ( 2003). Evolution by gene duplication: an update. Trend Ecol Evol 18, 292-298. |
| [36] | Zhu YG, Fu P, Lin QH, Zhang GT, Zhang HB, Li SM, Ju JH, Zhu WM, Zhang CS ( 2012). Identification of caerulomycin A gene cluster implicates a tailoring amidohydrolase. Org Lett 14, 2666-2669. |
| [37] | Zhu YG, Picard ME, Zhang QB, Barma J, Després XM, Mei XG, Zhang LP, Duvignaud JB, Couture M, Zhu WM, Shi R, Zhang CS ( 2016). Flavoenzyme CrmK-mediated substrate recycling in caerulomycin biosynthesis. Chem Sci 7, 4867-4874. |
| [1] | 刘雨函, 曹启江, 张诗晗, 李益慧, 王菁, 谭晓萌, 刘筱儒, 王显玲. 拟南芥AtFTCD-L参与根系响应土壤紧实度的机制研究[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 陈楠, 张全国. 实验进化研究途径[J]. 生物多样性, 2024, 32(9): 24171-. |
| [4] | 何花, 谭敦炎, 杨晓琛. 被子植物隐性雌雄异株性系统的多样性、系统演化及进化意义[J]. 生物多样性, 2024, 32(6): 24149-. |
| [5] | 景艳军, 林荣呈. 蓝光受体CRY2化身“暗黑舞者”[J]. 植物学报, 2024, 59(6): 878-882. |
| [6] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [7] | 曲锐, 左振君, 王有鑫, 张良键, 吴志刚, 乔秀娟, 王忠. 基于元素组的生物地球化学生态位及其在不同生态系统中的应用[J]. 生物多样性, 2024, 32(4): 23378-. |
| [8] | 杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础[J]. 植物学报, 2024, 59(3): 373-382. |
| [9] | 陈艳晓, 李亚萍, 周晋军, 解丽霞, 彭永彬, 孙伟, 和亚男, 蒋聪慧, 王增兰, 郑崇珂, 谢先芝. 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024, 59(3): 481-494. |
| [10] | 朱璐, 袁冲, 刘义飞. 植物次生代谢产物生物合成基因簇研究进展[J]. 植物学报, 2024, 59(1): 134-143. |
| [11] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [12] | 沈诗韵, 潘远飞, 陈丽茹, 土艳丽, 潘晓云. 喜旱莲子草原产地和入侵地种群的植物-土壤反馈差异[J]. 生物多样性, 2023, 31(3): 22436-. |
| [13] | 王钢, 王二涛. “卫青不败由天幸”——WeiTsing的广谱抗根肿病机理被揭示[J]. 植物学报, 2023, 58(3): 356-358. |
| [14] | 席念勋, 张原野, 周淑荣. 群落生态学中的植物-土壤反馈研究[J]. 植物生态学报, 2023, 47(2): 170-182. |
| [15] | 戚海迪, 张定海, 单立山, 陈国鹏, 张勃. 昆虫病原真菌感染昆虫宿主的机制和宿主昆虫的防御策略研究进展[J]. 生物多样性, 2023, 31(11): 23273-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||