

植物学报 ›› 2019, Vol. 54 ›› Issue (1): 93-101.DOI: 10.11983/CBB18044 cstr: 32102.14.CBB18044
魏铭1,2,王鑫伟1,陈博1,宋程威1,杜亮1,肖建伟1,林金星1,*( )
)
收稿日期:2018-02-09
接受日期:2018-05-23
出版日期:2019-01-01
发布日期:2019-07-31
通讯作者:
林金星
基金资助:
Ming Wei1,2,Xinwei Wang1,Bo Chen1,Chengwei Song1,Liang Du1,Jianwei Xiao1,Jinxing Lin1,*( )
)
Received:2018-02-09
Accepted:2018-05-23
Online:2019-01-01
Published:2019-07-31
Contact:
Jinxing Lin
摘要: 紫色酸性磷酸酶(PAPs)是一类广泛存在于植物体内的金属磷酸酯酶, 其羧基端含有1个保守结构域, 由5个保守基序和7个氨基酸残基构成。作为一种特殊的酸性磷酸酶, PAPs在酸性环境下能够有效催化磷酸酯或酸酐的水解, 释放出植物可以利用的磷酸基团。此外, PAPs在调节植物碳代谢、细胞壁合成和抵御病菌侵染等方面也发挥重要生理作用。该文简要介绍了PAPs的结构、家族成员及其调控因子, 并着重总结了近年来对PAPs生物学功能的研究进展, 为今后系统开展PAPs功能研究提供了理论参考。
魏铭,王鑫伟,陈博,宋程威,杜亮,肖建伟,林金星. 植物紫色酸性磷酸酶基因家族功能研究进展. 植物学报, 2019, 54(1): 93-101.
Ming Wei,Xinwei Wang,Bo Chen,Chengwei Song,Liang Du,Jianwei Xiao,Jinxing Lin. Research Progress into the Function of Purple Acid Phosphatase Gene Family in Plants. Chinese Bulletin of Botany, 2019, 54(1): 93-101.
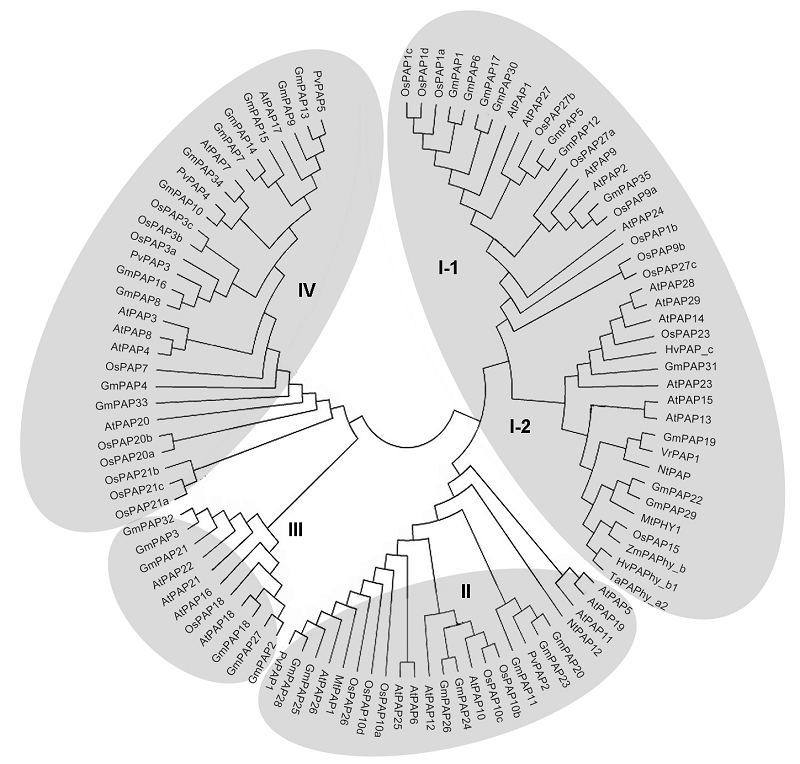
图2 植物PAPs物种间的进化关系(改自Tian and Liao, 2015) At: 拟南芥; Os: 水稻; Gm: 大豆; Mt: 蒺藜苜蓿; Nt: 烟草; Zm: 玉米; Hv: 大麦; Ta: 小麦; Vr: 绿豆; Pv: 菜豆
Figure 2 Phylogenetic relationships of plant PAPs (modified from Tian and Liao, 2015) At: Arabidopsis thaliana; Os: Oryza sativa; Gm: Glycine max; Mt: Medicago truncatula; Nt: Nicotiana tabacum; Zm: Zea mays; Hv: Hordeum vulgare; Ta: Triticum aestivum; Vr: Vigna radiata; Pv: Phaseolus vulgaris
| 蛋白 | 物种 | 功能 | 参考文献 |
|---|---|---|---|
| AtPAP2 | 拟南芥 | 碳代谢 | Sun et al., 2012a |
| AtPAP10 | 拟南芥 | 参与初生壁合成 | Kaida et al., 2003 |
| AtPAP12 | 拟南芥 | 磷代谢 | Wang et al., 2014 |
| AtPAP15 | 拟南芥 | 响应非生物逆境胁迫 | Zhang et al., 2008 |
| AtPAP25 | 拟南芥 | 磷代谢 | Del Vecchio et al., 2014 |
| AtPAP26 | 拟南芥 | 磷代谢 | Wang et al., 2014 |
| GmPAP3 | 大豆 | 响应非生物逆境胁迫 | Li et al., 2008 |
| GmPAP4 | 大豆 | 响应非生物逆境胁迫 | Kong et al., 2014 |
| NtPAP12 | 烟草 | 参与初生壁合成 | Kaida et al., 2010 |
| PvPAP3 | 菜豆 | 磷代谢 | Liang et al., 2010 |
| StPAP1 | 马铃薯 | 磷代谢 | Zimmermann et al., 2004 |
表1 植物PAP的生物学特性和功能(改自Tian and Liao, 2015)
Table 1 Biochemical properties functions of plant PAPs (modified from Tian and Liao, 2015)
| 蛋白 | 物种 | 功能 | 参考文献 |
|---|---|---|---|
| AtPAP2 | 拟南芥 | 碳代谢 | Sun et al., 2012a |
| AtPAP10 | 拟南芥 | 参与初生壁合成 | Kaida et al., 2003 |
| AtPAP12 | 拟南芥 | 磷代谢 | Wang et al., 2014 |
| AtPAP15 | 拟南芥 | 响应非生物逆境胁迫 | Zhang et al., 2008 |
| AtPAP25 | 拟南芥 | 磷代谢 | Del Vecchio et al., 2014 |
| AtPAP26 | 拟南芥 | 磷代谢 | Wang et al., 2014 |
| GmPAP3 | 大豆 | 响应非生物逆境胁迫 | Li et al., 2008 |
| GmPAP4 | 大豆 | 响应非生物逆境胁迫 | Kong et al., 2014 |
| NtPAP12 | 烟草 | 参与初生壁合成 | Kaida et al., 2010 |
| PvPAP3 | 菜豆 | 磷代谢 | Liang et al., 2010 |
| StPAP1 | 马铃薯 | 磷代谢 | Zimmermann et al., 2004 |
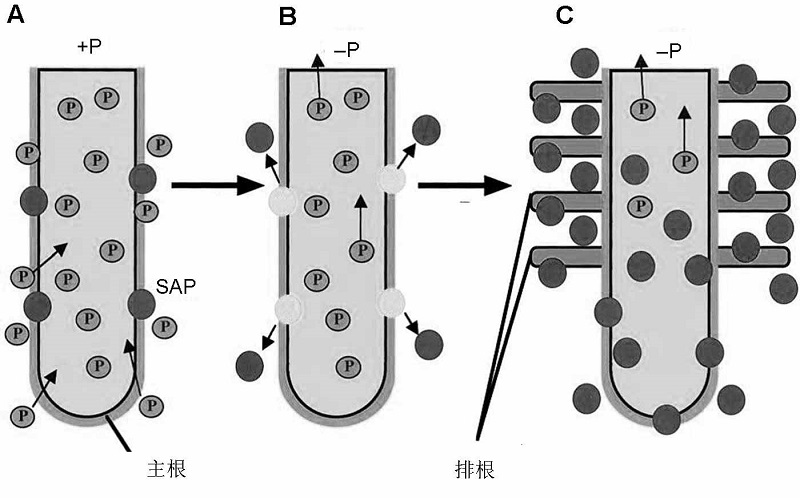
图3 缺磷情况下分泌型酸性磷酸酶的诱导表达及其根际分泌与排根形成示意图(改自Wasaki et al., 2003) (A) 磷充足条件下, SAP聚集在根表皮细胞周围, 只有少量SAP分泌; (B) 磷胁迫条件下, SAP在12小时内从根部迅速分泌, 但在长时间保持较低水平; (C) 组织中磷含量明显下降后, 根系成簇排列, 形成排根, SAP分泌量显著增加。
Figure 3 Schematic model of SAP expression and formation of cluster root under phosphorus deficiency (modified from Wasaki et al., 2003) (A) Under sufficient P conditions, SAP localizes around the epidermal cells of roots, and only a small amount of SAP is secreted; (B) After P stress treatment, SAP is released from roots rapidly within 12 h, however, SAP secretion remains at a low level for a long time; (C) After the decrease of P content in tissues, cluster roots form and SAP secretion significantly increases.
| 1 |
刘涛, 陈海英, 余海英, 李廷轩, 高尚卿, 陈光登 ( 2016). 低磷胁迫下大麦叶片磷素利用特征. 植物学报 51, 504-514.
DOI URL |
| 2 |
卢坤, 张凯, 柴友荣, 陆俊杏, 唐章林, 李加纳 ( 2010). 甘蓝和白菜紫色酸性磷酸酶17基因家族的克隆和比较分析. 作物学报 36, 517-525.
DOI URL |
| 3 |
周志高, 汪金舫, 周健民 ( 2005). 植物磷营养高效的分子生物学研究进展. 植物学报 22, 82-91.
DOI URL |
| 4 |
Cakmak I ( 2002). Plant nutrition research: priorities to meet human needs for food in sustainable ways. Plant Soil 247, 3-24.
DOI URL |
| 5 |
Del Vecchio HA, Ying S, Park J, Knowles VL, Kanno S, Tanoi K, She YM, Plaxton WC ( 2014). The cell wall- targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation.Plant J 80, 569-581.
DOI URL PMID |
| 6 |
Hammond JP, Broadley MR, White PJ ( 2004). Genetic responses to phosphorus deficiency. Ann Bot 94, 323-332.
DOI URL PMID |
| 7 |
Hur YJ, Jin BR, Nam J, Chung YS, Lee JH, Choi HK, Yun DJ, Yi G, Kim YH, Kim DH ( 2010). Molecular characterization of OsPAP2: transgenic expression of a purple acid phosphatase up-regulated in phosphate-deprived rice sus- pension cells.Biotechnol Lett 32, 163-170.
DOI URL PMID |
| 8 |
James J, David WL ( 1992). Dependence of photosynthesis of sunflower and maize leaves on phosphate supply, ribulose-1,5-bisphosphate carboxylase/oxygenase activity, and ribulose-1,5-bisphosphate pool size. Plant Physiol 98, 801-807.
DOI URL PMID |
| 9 |
Jarenmark M, Haukka M, Demeshko S, Tuczek F, Zuppiroli L, Meyer F, Nordlander E ( 2011). Synthesis, characterization, and reactivity studies of heterodinuclear complexes modeling active sites in purple acid phospha- tases. Inorg Chem 50, 3866-3887.
DOI URL PMID |
| 10 |
Kaida R, Hayashi T, Kaneko TS ( 2008). Purple acid phosphatase in the walls of tobacco cells. Phytochemistry 69, 2546-2551.
DOI URL PMID |
| 11 |
Kaida R, Sage-Ono K, Kamada H, Okuyama H, Syono K, Kaneko TS ( 2003). Isolation and characterization of four cell wall purple acid phosphatase genes from tobacco cells. Biochim Biophys Acta-Gene Regul Mech 1625, 134-140.
DOI URL PMID |
| 12 |
Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS ( 2009). Activation of β-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol 150, 1822-1830.
DOI URL PMID |
| 13 |
Kaida R, Serada S, Norioka N, Norioka S, Neumetzler L, Pauly M, Sampedro J, Zarra I, Hayashi T, Kaneko TS ( 2010). Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol 153, 603-610.
DOI URL PMID |
| 14 |
Klabunde T, Stahl B, Suerbaum H, Hahner S, Karas M, Hillenkamp F, Krebs B, Witzel H ( 1994). The amino acid sequence of the red kidney bean Fe(III)-Zn(II) purple acid phosphatase. Determination of the amino acid sequence by a combination of matrix-assisted laser desorption/ioni- zation mass spectrometry and automated Edman sequ- encing. Eur J Biochem 226, 369-375.
DOI URL PMID |
| 15 |
Klabunde T, Sträter N, Krebs B, Witzel H ( 1995). Structural relationship between the mammalian Fe(III)-Fe(II) and the Fe(III)-Zn(II) plant purple acid phosphatases. FEBS Lett 367, 56-60.
DOI URL PMID |
| 16 | Kong YB, Li XH, Ma J, Li WL, Yan GJ, Zhang CY ( 2014). GmPAP4, a novel purple acid phosphatase gene isolated from soybean ( Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana.Plant Cell Rep 33, 655-667. |
| 17 |
Kuang RB, Chan KH, Yeung E, Lim BL ( 2009). Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol 151, 199-209.
DOI URL PMID |
| 18 |
Kusudo T, Sakaki T, Inouye K ( 2003). Purification and characterization of purple acid phosphatase PAP1 from dry powder of sweet potato. Biosci Biotechnol Biochem 67, 1609-1611.
DOI URL |
| 19 |
Law YS, Zhang RS, Guan XQ, Cheng SF, Sun F, Duncan O, Murcha MW, Whelan J, Lim BL ( 2015). Phosphorylation and dephosphorylation of the presequence of precursor MULTIPLE ORGANELLAR RNA EDITING FACTOR3 during import into mitochondria from Arabidopsis. Plant Physiol 169, 1344-1355.
DOI URL PMID |
| 20 |
Li CC, Gui SH, Yang T, Walk T, Wang XR, Liao H ( 2012). Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann Bot 109, 275-285.
DOI URL PMID |
| 21 |
Li DP, Zhu HF, Liu KF, Liu X, Leggewie G, Udvardi M, Wang DW ( 2002). Purple acid phosphatases of Arabidopsis thaliana.Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277, 27772-27781.
DOI URL PMID |
| 22 |
Li WYF, Shao GH, Lam HM ( 2008). Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses.New Phytol 178, 80-91.
DOI URL PMID |
| 23 |
Liang C, Liu X, Sun YZ, Yiu SM, Lim BL ( 2014). Global small RNA analysis in fast-growing Arabidopsis thaliana with elevated concentrations of ATP and sugars.BMC Genomics 15, 116.
DOI URL PMID |
| 24 |
Liang C, Zhang YJ, Cheng SF, Osorio S, Sun YZ, Fernie AR, Cheung CY, Lim BL ( 2015). Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana.Front Plant Sci 6, 922.
DOI URL PMID |
| 25 |
Liang CY, Tian J, Lam HM, Lim BL, Yan XL, Liao H ( 2010). Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152, 854-865.
DOI URL |
| 26 |
Liao H, Wong FL, Phang TH, Cheung MY, Li WYF, Shao GH, Yan XL, Lam HM ( 2003). GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318, 103-111.
DOI URL PMID |
| 27 | Nakazato H, Okamoto T, Nishikoori M, Washio K, Morita N, Haraguchi K, Thompson GA, Okuyama H ( 1998). The glycosylphosphatidylinositol-anchored phosphatase from Spirodela oligorrhiza is a purple acid phosphatase.Plant Physiol 118, 1015-1020. |
| 28 |
Olczak M, Morawiecka B, Watorek W ( 2003). Plant purple acid phosphatases-genes, structures and biological function. Acta Biochim Pol 50, 1245-1256.
DOI URL PMID |
| 29 | Ravichandran S, Stone SL, Bernhard B, Prithiviraj B ( 2013). Purple acid phosphatase 5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis.BMC Plant Biol 13, 107. |
| 30 |
Robinson WD, Carson I, Ying S, Ellis K, Plaxton WC ( 2012a). Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization.New Phytol 196, 1024-1029.
DOI URL PMID |
| 31 |
Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC ( 2012 b). The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana.J Exp Bot 63, 6531-6542.
DOI URL PMID |
| 32 |
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J ( 2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15, 2122-2133.
DOI URL |
| 33 | Schenk G, Elliott TW, Leung E, Carrington LE, Mitic N, Gahan LR, Guddat LW ( 2008). Crystal structures of a purple acid phosphatase, representing different steps of this enzyme’s catalytic cycle. BMC Struct Biol 8, 6. |
| 34 |
Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J ( 2000 a). Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250, 117-125.
DOI URL PMID |
| 35 |
Schenk G, Korsinczky MLJ, Hume DA, Hamilton S, DeJersey J ( 2000 b). Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes. Gene 255, 419-424.
DOI URL PMID |
| 36 |
Smith VH, Schindler DW ( 2009). Eutrophication science: where do we go from here? Trends Ecol Evol 24, 201-207.
DOI URL PMID |
| 37 | Strater N, Klabunde T, Tucker P, Witzel H, Krebs B ( 1995). Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science 268, 1489-1492. |
| 38 |
Sun F, Carrie C, Law S, Murcha MW, Zhang R, Law YS, Suen PK, Whelan J, Lim BL ( 2012 b). AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal Behav 7, 927-932.
DOI URL PMID |
| 39 | Sun F, Liang C, Whelan J, Yang J, Zhang P, Lim BL ( 2013). Global transcriptome analysis of AtPAP2-overex- pressing Arabidopsis thaliana with elevated ATP.BMC Genomics 14, 752. |
| 40 |
Sun F, Suen PK, Zhang YJ, Liang C, Carrie C, Whelan J, Ward JL, Hawkins ND, Jiang LW, Lim BL ( 2012a). A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield.New Phytol 194, 206-219.
DOI URL PMID |
| 41 |
Sun YZ, Law YS, Cheng SF, Lim BL ( 2017). RNA editing of cytochrome c maturation transcripts is responsive to the energy status of leaf cells in Arabidopsis thaliana.Mitochondrion 35, 23-34.
DOI URL PMID |
| 42 |
Tian J, Liao H (2015). The role of intracellular and secreted purple acid phosphatases in plant phosphorus scavenging and recycling. In: Plaxton WC, Lambers H, eds. Annual Plant Reviews Volume 48: Phosphorus Metabolism in Plants, Vol. 48. West Sussex: John Wiley & Sons, Ltd,. Ltd. pp. 265-287.
DOI URL |
| 43 |
Tran HT, Hurley BA, Plaxton WC ( 2010 a). Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179, 14-27.
DOI URL |
| 44 | Tran HT, Qian WQ, Hurley BA, She YM, Wang DW, Plaxton WC ( 2010 b). Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate- starved Arabidopsis thaliana.Plant Cell Environ 33, 1789-1803. |
| 45 |
Vance CP ( 2001). Symbiotic nitrogen fixation and phosphorus acquisition. plant nutrition in a world of declining renewable resources. Plant Physiol 127, 390-397.
DOI URL PMID |
| 46 |
Vance CP, Uhde-Stone C, Allan DL ( 2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157, 423-447.
DOI URL |
| 47 |
Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, Raven JA ( 2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol 195, 306-320.
DOI URL PMID |
| 48 | Wang LS, Li Z, Qian WQ, Guo WL, Gao X, Huang LL, Wang H, Zhu HF, Wu JW, Wang DW, Liu D ( 2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157, 1283-1299. |
| 49 | Wang LS, Lu S, Zhang Y, Li Z, Du XQ, Liu D ( 2014). Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56, 299-314. |
| 50 |
Wasaki J, Yamamura T, Shinano T, Osaki M ( 2003). Secreted acid phosphatase is expressed in cluster roots of lupin in response to phosphorus deficiency. Plant Soil 248, 129-136.
DOI URL |
| 51 |
Zhang Q, Wang C, Tian J, Li K, Shou H ( 2011). Identification of rice purple acid phosphatases related to phosphate starvation signaling. Plant Biol 13, 7-15.
DOI URL PMID |
| 52 |
Zhang RS, Guan XQ, Law YS, Sun F, Chen S, Wong KB, Lim BL ( 2016). AtPAP2 modulates the import of the small subunit of Rubisco into chloroplasts. Plant Signal Behav 11, e1239687.
DOI URL PMID |
| 53 |
Zhang WY, Gruszewski HA, Chevone BI, Nessler CL ( 2008). An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146, 431-440.
DOI URL PMID |
| 54 |
Zhang YJ, Sun F, Fettke J, Schottler MA, Ramsden L, Fernie AR, Lim BL ( 2014). Heterologous expression of AtPAP2 in transgenic potato influences carbon metabolism and tuber development.FEBS Lett 588, 3726-3731.
DOI URL PMID |
| 55 |
Zhang YJ, Yu L, Yung KF, Leung DYC, Sun F, Lim BL ( 2012). Over-expression of AtPAP2 in Camelina sativa leads to faster plant growth and higher seed yield.Biotechnol Biofuels 5, 219.
DOI URL PMID |
| 56 |
Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He XW, Zhong WQ, Wu P ( 2008). OsPHR2 is involved in phosphate- starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146, 1673-1686.
DOI URL PMID |
| 57 |
Zhu HF, Qian WQ, Lu XZ, Li DP, Liu X, Liu KF, Wang DW ( 2005). Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower.Plant Mol Biol 59, 581-594.
DOI URL PMID |
| 58 |
Zimmermann P, Regierer B, Kossmann J, Frossard E, Amrhein N, Bucher M ( 2004). Differential expression of three purple acid phosphatases from potato. Plant Biol 6, 519-528.
DOI URL PMID |
| [1] | 王子韵, 吕燕文, 肖钰, 吴超, 胡新生. 植物基因表达调控与进化机制研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 胡海涛, 武越, 杨玲. 植物NAD(P)+的生物合成及其生物学功能研究进展[J]. 植物学报, 2025, 60(1): 114-131. |
| [3] | 李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41. |
| [4] | 贺祯媚,李东明,齐艳华. 植物ABCB亚家族生物学功能研究进展[J]. 植物学报, 2019, 54(6): 688-698. |
| [5] | 胡佳, 刘春林. 植物油体研究进展[J]. 植物学报, 2017, 52(5): 669-679. |
| [6] | 席红梅, 徐文忠, 麻密. 拟南芥双功能酶SAL1生物学功能的研究进展[J]. 植物学报, 2016, 51(3): 377-386. |
| [7] | 俞乐, 刘拥海, 袁伟超, 周丽萍, 彭长连. 植物抗坏血酸积累及其分子机制的研究进展[J]. 植物学报, 2016, 51(3): 396-410. |
| [8] | 刘林娅, 黄亚成, 黄小龙, 黄东益. 薯蓣植物块茎特异蛋白Dioscorin的研究进展[J]. 植物学报, 2016, 51(2): 274-280. |
| [9] | 刘海娇, 杜立群, 林金星, 李瑞丽. 植物环核苷酸门控离子通道及其功能研究进展[J]. 植物学报, 2015, 50(6): 779-789. |
| [10] | 翟开恩, 潘伟槐, 叶晓帆, 潘建伟. 高等植物局部生长素合成的生物学功能及其调控机制[J]. 植物学报, 2015, 50(2): 149-158. |
| [11] | 李明, 李长生, 赵传志, 李爱芹, 王兴军. 植物SPL转录因子研究进展[J]. 植物学报, 2013, 48(1): 107-116. |
| [12] | 刘润华;江文波;余迪求. 植物鞘脂的结构、代谢途径及其功能[J]. 植物学报, 2009, 44(05): 619-628. |
| [13] | 唐中华 于景华 杨逢建 祖元刚. 植物生物碱代谢生物学研究进展[J]. 植物学报, 2003, 20(06): 696-702. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||