

植物学报 ›› 2018, Vol. 53 ›› Issue (5): 726-737.DOI: 10.11983/CBB17078 cstr: 32102.14.CBB17078
• 专题论坛 • 上一篇
收稿日期:2017-04-09
接受日期:2017-08-30
出版日期:2018-09-01
发布日期:2018-11-29
通讯作者:
陈国雄
作者简介:† 共同第一作者。
基金资助:
Zhang Jiwei1,2, Zhao Jiecai1, Zhou Qin1,2, Chen Guoxiong1,*( )
)
Received:2017-04-09
Accepted:2017-08-30
Online:2018-09-01
Published:2018-11-29
Contact:
Chen Guoxiong
About author:† These authors contributed equally to this paper
摘要: 表皮毛是大多数植物地上部分表皮组织所延伸出来的一种特化的毛状结构附属物。表皮毛在植物表皮层和环境间构筑了一道天然的物理屏障, 不但对植物的生长发育具有重要意义, 而且还具有非常高的应用价值和经济价值。近几年, 研究者从不同植物中不断克隆出新的表皮毛发育相关基因, 在揭示植物调控表皮毛生长发育的分子机制方面取得很大进展。该文综述了植物表皮毛的最新研究进展, 并展望了植物表皮毛的研究方向及应用开发价值。
张继伟, 赵杰才, 周琴, 陈国雄. 植物表皮毛研究进展. 植物学报, 2018, 53(5): 726-737.
Zhang Jiwei, Zhao Jiecai, Zhou Qin, Chen Guoxiong. Progress in Research of Plant Trichome. Chinese Bulletin of Botany, 2018, 53(5): 726-737.
| 基因名称 | 物种 | 基因序列号 | 产物 | 功能及突变体表型描述 | 参考文献 |
|---|---|---|---|---|---|
| GL1 | 拟南芥 | AF495524 | R2R3-MYB类型转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛, 但过表达反而导致表皮毛减少 | |
| TTG1 | 拟南芥 | BT011856 | 含有WD40重复序列蛋白 | 调控表皮毛的启动, 突变体表现为无表皮毛, 且突变体种子的休眠特性消失 | |
| GL3 | 拟南芥 | AF246291 | bHLH类型转录因子 | 调控表皮毛的启动, 突变体表现为表皮毛数量减少 | |
| GL2 | 拟南芥 | AJ457046 | HD2-ZIP结构转录因子 | 调控表皮细胞命运, 突变体表皮毛减少, 且大部分无分枝 | |
| TTG2 | 拟南芥 | AF404862 | WRKY类型转录因子 | 控制表皮毛早期发育, 突变体表现为分枝减少或无分枝 | |
| EGL3 | 拟南芥 | AF251687 | bHLH类型转录因子 | 正调节因子, 功能与GL3冗余, 而且gl3/egl3双突变体表现为无表皮毛 | |
| MYB23 | 拟南芥 | Z68158 | R2R3-MYB类型转录因子 | 异位表达可以促进表皮毛生长 | |
| TRY | 拟南芥 | AY161286 | R3-MYB类型转录因子 | 负调节因子, 突变体表皮毛成簇生长且分枝增加 | |
| CPC | 拟南芥 | AB004871 | R3-MYB类型转录因子 | 负调节因子, 突变体表现为根毛减少, 表皮毛簇生 | |
| WER | 拟南芥 | Z95789 | R2R3-MYB类型转录因子 | 控制根毛发育, 功能缺失导致根毛显著增加 | |
| ETC1 | 拟南芥 | AY519518 | MYB类型转录因子 | 负调节因子 | |
| ETC2 | 拟南芥 | AY234411 | MYB类型转录因子 | 负调节因子 | |
| ETC3 | 拟南芥 | AB264292 | MYB类型转录因子 | 突变体表现为表皮毛分枝增加而根毛密度减少 | |
| KAK | 拟南芥 | AY265959 | 泛素连接酶 | 调节表皮毛核内复制, 突变体表现为表皮毛分枝增加, DNA含量增加 | |
| GIS | 拟南芥 | BT003340 | 含C2H2的转录因子 | 调控表皮细胞和花序的分化, 突变体表皮毛减少 | |
| GIS2 | 拟南芥 | NM_120748 | 含C2H2的转录因子 | 调控表皮毛的分化, 受赤霉素和细胞分裂素的调节 | |
| RHL2 | 拟南芥 | AJ297842 | DNA拓扑异构酶 | 调节表皮毛核内复制, 调控表皮毛形态 | |
| STI | 拟南芥 | AF264023 | 未知 | 调控表皮毛分枝形成, 突变体表现为表皮毛分枝减少 | |
| KLK | 拟南芥 | AY496701 | 未知 | 调控表皮毛分枝形成 | |
| BRICK1 | 拟南芥 | AY085644 | 未知 | 调控表皮毛分枝形成 | |
| DIS1 | 拟南芥 | AF507911 | 未知 | 调控表皮毛分枝形成, 突变体表现为表皮毛细胞扩增, 且形态不正常 | |
| DIS2 | 拟南芥 | AY084256 | 未知 | 调控表皮毛分枝形成 | |
| SPL9 | 拟南芥 | AJ011638 | 未知 | 受miR156调控, 抑制表皮毛的启动 | |
| TEM | 拟南芥 | AY056169 | RAV转录因子 | 光周期响应因子, 参与调控赤霉素合成与转运, 调节花期与表皮毛形成等 | |
| TCS1 | 拟南芥 | AK222122 | 未知 | 突变体表现为表皮毛有缺陷 | |
| 基因名称 | 物种 | 基因序列号 | 产物 | 功能及突变体表型描述 | 参考文献 |
| TRIL | 黄瓜 | 未知 | HD-ZIP结构转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛 | |
| CsGL1 | 黄瓜 | 未知 | HD-ZIP结构转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛 | |
| GoPGF | 棉花 | KP072743 | bHLH类型转录因子 | 调控表皮毛的启动, 沉默GoPGF表现为无表皮毛 | |
| GhMYB2A | 棉花 | JQ868556 | MYB类型转录因子 | 调控棉花纤维形成, 受miRNA调节 | |
| GhMYB2D | 棉花 | JQ868560 | MYB类型转录因子 | 调控棉花纤维形成, 受miRNA调节 | |
| AaMYB1 | 黄花蒿 | KC118530 | MYB类型转录因子 | 调控表皮毛发育和萜类代谢 | |
| AaHD1 | 黄花蒿 | KU744599 | HD-ZIP结构转录因子 | 调控表皮毛发育, 过表达导致表皮毛密度增加 | |
| HI | 番茄 | 未知 | SRA1的1个亚基 | 调控肌动蛋白丝凝聚, 突变体表皮毛形态有缺陷 |
表1 植物表皮毛发育相关重要基因
Table 1 The important genes related to trichome development in plants
| 基因名称 | 物种 | 基因序列号 | 产物 | 功能及突变体表型描述 | 参考文献 |
|---|---|---|---|---|---|
| GL1 | 拟南芥 | AF495524 | R2R3-MYB类型转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛, 但过表达反而导致表皮毛减少 | |
| TTG1 | 拟南芥 | BT011856 | 含有WD40重复序列蛋白 | 调控表皮毛的启动, 突变体表现为无表皮毛, 且突变体种子的休眠特性消失 | |
| GL3 | 拟南芥 | AF246291 | bHLH类型转录因子 | 调控表皮毛的启动, 突变体表现为表皮毛数量减少 | |
| GL2 | 拟南芥 | AJ457046 | HD2-ZIP结构转录因子 | 调控表皮细胞命运, 突变体表皮毛减少, 且大部分无分枝 | |
| TTG2 | 拟南芥 | AF404862 | WRKY类型转录因子 | 控制表皮毛早期发育, 突变体表现为分枝减少或无分枝 | |
| EGL3 | 拟南芥 | AF251687 | bHLH类型转录因子 | 正调节因子, 功能与GL3冗余, 而且gl3/egl3双突变体表现为无表皮毛 | |
| MYB23 | 拟南芥 | Z68158 | R2R3-MYB类型转录因子 | 异位表达可以促进表皮毛生长 | |
| TRY | 拟南芥 | AY161286 | R3-MYB类型转录因子 | 负调节因子, 突变体表皮毛成簇生长且分枝增加 | |
| CPC | 拟南芥 | AB004871 | R3-MYB类型转录因子 | 负调节因子, 突变体表现为根毛减少, 表皮毛簇生 | |
| WER | 拟南芥 | Z95789 | R2R3-MYB类型转录因子 | 控制根毛发育, 功能缺失导致根毛显著增加 | |
| ETC1 | 拟南芥 | AY519518 | MYB类型转录因子 | 负调节因子 | |
| ETC2 | 拟南芥 | AY234411 | MYB类型转录因子 | 负调节因子 | |
| ETC3 | 拟南芥 | AB264292 | MYB类型转录因子 | 突变体表现为表皮毛分枝增加而根毛密度减少 | |
| KAK | 拟南芥 | AY265959 | 泛素连接酶 | 调节表皮毛核内复制, 突变体表现为表皮毛分枝增加, DNA含量增加 | |
| GIS | 拟南芥 | BT003340 | 含C2H2的转录因子 | 调控表皮细胞和花序的分化, 突变体表皮毛减少 | |
| GIS2 | 拟南芥 | NM_120748 | 含C2H2的转录因子 | 调控表皮毛的分化, 受赤霉素和细胞分裂素的调节 | |
| RHL2 | 拟南芥 | AJ297842 | DNA拓扑异构酶 | 调节表皮毛核内复制, 调控表皮毛形态 | |
| STI | 拟南芥 | AF264023 | 未知 | 调控表皮毛分枝形成, 突变体表现为表皮毛分枝减少 | |
| KLK | 拟南芥 | AY496701 | 未知 | 调控表皮毛分枝形成 | |
| BRICK1 | 拟南芥 | AY085644 | 未知 | 调控表皮毛分枝形成 | |
| DIS1 | 拟南芥 | AF507911 | 未知 | 调控表皮毛分枝形成, 突变体表现为表皮毛细胞扩增, 且形态不正常 | |
| DIS2 | 拟南芥 | AY084256 | 未知 | 调控表皮毛分枝形成 | |
| SPL9 | 拟南芥 | AJ011638 | 未知 | 受miR156调控, 抑制表皮毛的启动 | |
| TEM | 拟南芥 | AY056169 | RAV转录因子 | 光周期响应因子, 参与调控赤霉素合成与转运, 调节花期与表皮毛形成等 | |
| TCS1 | 拟南芥 | AK222122 | 未知 | 突变体表现为表皮毛有缺陷 | |
| 基因名称 | 物种 | 基因序列号 | 产物 | 功能及突变体表型描述 | 参考文献 |
| TRIL | 黄瓜 | 未知 | HD-ZIP结构转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛 | |
| CsGL1 | 黄瓜 | 未知 | HD-ZIP结构转录因子 | 调控表皮毛的启动, 突变体表现为无表皮毛 | |
| GoPGF | 棉花 | KP072743 | bHLH类型转录因子 | 调控表皮毛的启动, 沉默GoPGF表现为无表皮毛 | |
| GhMYB2A | 棉花 | JQ868556 | MYB类型转录因子 | 调控棉花纤维形成, 受miRNA调节 | |
| GhMYB2D | 棉花 | JQ868560 | MYB类型转录因子 | 调控棉花纤维形成, 受miRNA调节 | |
| AaMYB1 | 黄花蒿 | KC118530 | MYB类型转录因子 | 调控表皮毛发育和萜类代谢 | |
| AaHD1 | 黄花蒿 | KU744599 | HD-ZIP结构转录因子 | 调控表皮毛发育, 过表达导致表皮毛密度增加 | |
| HI | 番茄 | 未知 | SRA1的1个亚基 | 调控肌动蛋白丝凝聚, 突变体表皮毛形态有缺陷 |
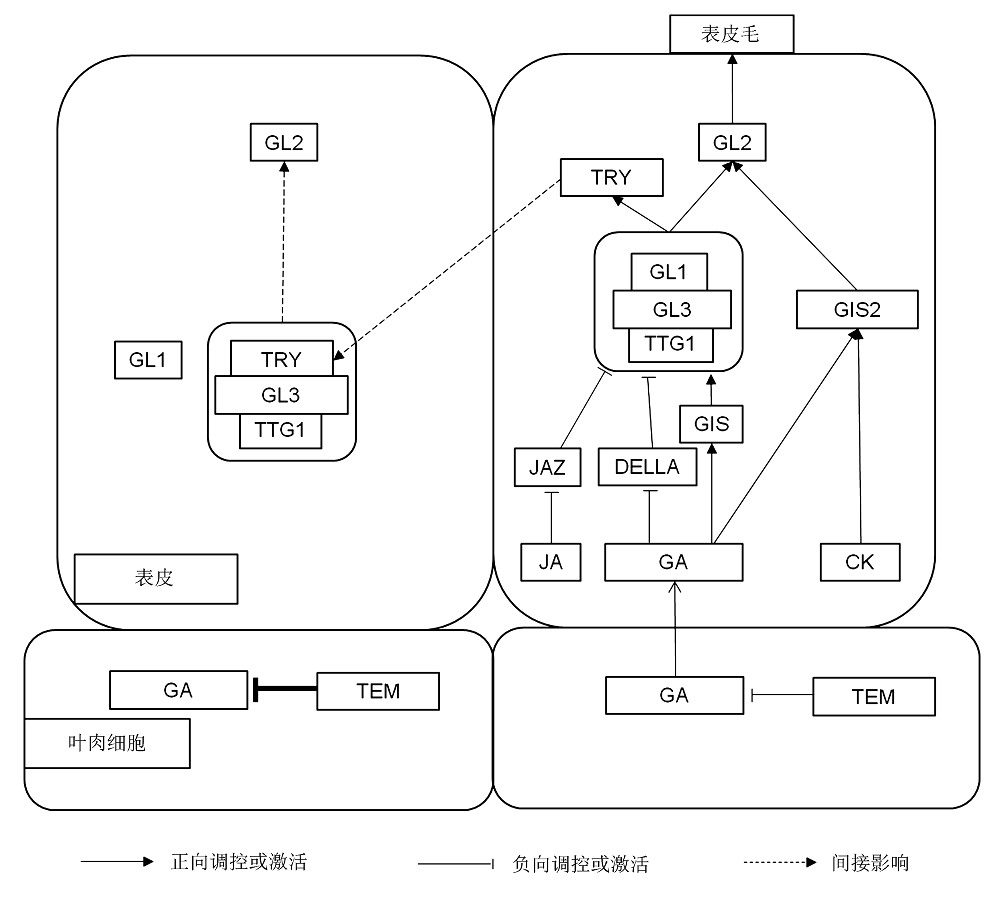
图1 植物表皮毛分化调控模式该模式显示, 赤霉素在叶肉细胞中的合成及转运是由TEM负责调控, 并在表皮毛前体细胞下面的叶肉细胞中积累和向上转运。赤霉素通过DELLA和GIS调控GL1-GL3-TTG1复合体, 茉莉酸通过JAZ也调控该复合体, 赤霉素和细胞分裂素调节GIS2, 然后, GL1-GL3-TTG1复合体和GIS2进一步调节GL2, 指导表皮毛发育。TRY等负调节因子移动至邻近表皮细胞, 与GL1竞争性结合到GL3上, 阻碍GL2的表达, 从而抑制表皮毛形成。
Figure 1 The regulate pattern of plant trichome differentiationThe diagram shows the synthesis and transport of GA in mesophyll are regulated by TEM, which also accumulate in mesophyll underneath pre-trichome and transfer to trichome. GA regulates the GL1-GL3-TTG1 complex through DELLA and GIS, JA also regulates the complex through JAZ, GA and CK regulate GIS2, then the complex and GIS2 guide trichome development by regulating GL2. The negative regulator TRY moves to neighboring epidermis where it competes with GL1 for binding to GL3, which suppresses the expression of GL2 and inhibit the trichome formation.
| 3 | 刘金秋, 陈凯, 张珍珠, 陈秀玲, 王傲雪 (2016). 外施GA、MeJA、IAA、SA和KT对番茄表皮毛发生的作用. 园艺学报 43, 2151-2160. |
| 4 | 卢阳, 龙鸿 (2015). 拟南芥叶片数目变化突变体对营养生长时相转变的影响. 植物学报 50, 331-337. |
| 5 | 马亚丽, 王璐, 刘艳霞, 兰海燕 (2015). 荒漠植物几种主要附属结构的抗逆功能及其协同调控的研究进展. 植物生理学报 51, 1821-1836. |
| 6 | 王翠丽, 田海霞, 汪姣姣, 齐天从, 黄煌, 任春梅, 谢道昕, 宋素胜 (2014). 拟南芥转录因子bHLH17负调控茉莉素介导的抗性反应. 植物学报 49, 643-652. |
| 7 | 杨淑华, 王台, 钱前, 王小菁, 左建儒, 顾红雅, 姜里文, 陈之端, 白永飞, 孔宏智, 陈凡, 萧浪涛, 董爱武, 种康 (2016). 2015年中国植物科学若干领域重要研究进展. 植物学报 51, 416-472. |
| 8 | Akhtar MQ, Qamar N, Yadav P, Kulkarni P, Kumar A, Shasany AK (2017). Comparative glandular trichome transcriptome-based gene characterization reveals reasons for differential (-)-menthol biosynthesis inMentha species. Physiol Plantarum 160, 128-141. |
| 9 | Alahakoon UI, Taheri A, Nayidu NK, Epp D, Yu M, Parkin I, Hegedus D, Bonham-Smith P, Gruber MY (2016). Hairy canola (Brasssica napus) re-visited: down-regulating TTG1 in an AtGL3-enhanced hairy leaf background improves growth, leaf trichome coverage, and metabolite gene expression diversity. BMC Plant Biol 16, 12. |
| 10 | Andrade MC, Da Silva AA, Neiva IP, Oliveira IRC, De Castro EM, Francis DM, Maluf WR (2017). Inheritance of type IV glandular trichome density and its association with whitefly resistance from Solanum galapagense accession LA1401. Euphytica 213, 52. |
| 11 | Atalay Z, Celep F, Bara F, Doğan M (2016). Systematic significance of anatomy and trichome morphology in Lamium(Lamioideae; Lamiaceae). Flora Morphol Distrib Funct Ecol Plants 225, 60-75. |
| 12 | Balkunde R, Pesch M, Hülskamp M (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol 91, 299-321. |
| 13 | Bensussan M, Lefebvre V, Ducamp A, Trouverie J, Emilie G, Fortabat MN, Guillebaux A, Baldy A, Naquin D, Herbette S (2015). Suppression of dwarf and irregular xylem phenotypes generates low-acetylated biomass lines in Arabidopsis. Plant Physiol 168, 452-463. |
| 14 | Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, Persson S, Delmer D, Scheible WR (2010). TRICHOME BIREFRINGENCE and its homolog AT5G- 01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol 153, 590-602. |
| 15 | Bryant L, Patole C, Cramer R (2016). Proteomic analysis of the medicinal plant Artemisia annua: data from leaf and trichome extracts. Data Brief 7, 325-331. |
| 16 | Busta L, Hegebarth D, Kroc E, Jetter R (2017). Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density.Planta 245, 297-311. |
| 17 | Champagne A, Boutry M (2017). A comprehensive proteome map of glandular trichomes of hop (Humulus lupulus L.) female cones: identification of biosynthetic pathways of the major terpenoid-related compounds and possible transport proteins. Proteomics 17, 1600411. |
| 18 | Chen GX, Zhao JC, Zhao X, Zhao PS, Duan RJ, Nevo E, Ma XF (2014). A psammophyte Agriophyllum squarrosum(L.) Moq: a potential food crop. Genet Resour Crop Evol 61, 669-676. |
| 19 | Chen LL, Peng YC, Tian J, Wang XH, Kong ZS, Mao TL, Yuan M, Li YH (2016). TCS1, a microtubule-binding protein, interacts with KCBP/ZWICHEL to regulate trichome cell shape in Arabidopsis thaliana. PLoS Genet 12, e100-6266. |
| 20 | Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016). Redundancy and specificity in jasmonate signaling.Curr Opin Plant Biol 33, 147-156. |
| 21 | Cui JY, Miao H, Ding LH, Wehner TC, Liu PN, Wang Y, Zhang SP, Gu XF (2016). A new glabrous gene (csgl3) identified in trichome development in cucumber(Cucumis sativus L.). PLoS One 11, e0148422. |
| 22 | Dai XB, Wang GD, Yang DS, Tang YH, Broun P, Marks MD, Sumner LW, Dixon RA, Zhao PX (2010). TrichOME: a comparative omics database for plant trichomes.Plant Physiol 152, 44-54. |
| 23 | Dai XM, Zhou LM, Zhang W, Cai L, Guo HY, Tian HN, Schiefelbein J, Wang SC (2016). A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3.Plant Cell Environ 39, 897-907. |
| 24 | Dyachok J, Zhu L, Liao FQ, He J, Huq E, Blancaflor EB (2011). SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes.Plant Cell 23, 3610-3626. |
| 25 | Gan YB, Liu C, Yu H, Broun P (2007). Integration of cytokinin and gibberellin signaling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate.Development 134, 2073-2081. |
| 26 | Gao CH, Li D, Jin CY, Duan SW, Qi SH, Liu KG, Wang HC, Ma HL, Hai JB, Chen MX (2017a). Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis.Biochem Biophys Res Commun 485, 360-365. |
| 27 | Gao YP, He CW, Zhang DM, Liu XL, Xu ZP, Tian YB, Liu XH, Zang SS, Pauly M, Zhou YH, Zhang BC (2017b). Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice.Plant Physiol 173, 470-481. |
| 28 | Guan XY, Pang MX, Nah G, Shi XL, Ye WX, Stelly DM, Chen ZJ (2014). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5, 3050. |
| 1 | 高英, 郭建强, 赵金凤 (2011). 拟南芥表皮毛发育的分子机制. 植物学报 46, 119-127. |
| 2 | 李根源, 郭香墨 (1992). 棉属植物表皮毛性状研究进展. 中国棉花 (6), 5-8. |
| 29 | Hegebarth D, Buschhaus C, Wu M, Bird D, Jetter R (2016). The composition of surface wax on trichomes of Arabidopsis thaliana differs from wax on other epidermal cells. Plant J 88, 762-774. |
| 30 | Hendrick MF, Finseth FR, Mathiasson ME, Palmer KA, Broder EM, Breigenzer P, Fishman L (2016). The genetics of extreme microgeographic adaptation: an integrated approach identifies a major gene underlying leaf trichome divergence in Yellowstone Mimulus guttatus. Mol Ecol 25, 5647-5662. |
| 31 | Inthima P, Nakano M, Otani M, Niki T, Nishijima T, Koshioka M, Supaibulwatana K (2017). Overexpression of the gibberellin 20-oxidase gene from Torenia fournieri resulted in modified trichome formation and terpenoid metabolities of Artemisia annua L. Plant Cell Tiss Organ Cult 129, 223-236. |
| 32 | Ioannidi E, Rigas S, Tsitsekian D, Daras G, Alatzas A, Makris A, Tanou G, Argiriou A, Alexandrou D, Poethig S, Hatzopoulos P, Kanellis AK (2016). Trichome patterning control involves TTG1 interaction with SPL transcription factors.Plant Mol Biol 92, 675-687. |
| 33 | Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19, 2531-2543. |
| 34 | Jiao YL (2016). Trichome formation: gibberellins on the move.Plant Physiol 170, 1174-1175. |
| 35 | Johnson CS, Kolevski B, Smyth DR (2002). TRANSPARENT TESTA GLABRA 2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359-1375. |
| 36 | Kang JH, Campos ML, Zemelis-Durfee S, Al-Haddad JM, Jones AD, Telewski FW, Brandizzi F, Howe GA (2016). Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J Exp Bot 67, 5313-5324. |
| 37 | Kang JH, Liu GH, Shi F, Jones AD, Beaudry RM, Howe GA (2010a). The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol 154, 262-272. |
| 38 | Kang JH, Shi F, Jones AD, Marks MD, Howe GA (2010b). Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61, 1053-1064. |
| 39 | Kariyat RR, Smith JD, Stephenson AG, De Moraes CM, Mescher MC (2017). Non-glandular trichomes of Solanum carolinense deter feeding by Manduca sexta caterpillars and cause damage to the gut peritrophic matrix. Proc Roy Soc B 284, 20162323. |
| 40 | Karray-Bouraoui N, Rabhi M, Neffati M, Baldan B, Ranieri A, Marzouk B, Lachaâl M, Smaoui A (2009). Salt effect on yield and composition of shoot essential oil and trichome morphology and density on leaves of Mentha pulegium. Ind Crops Prod 30, 338-343. |
| 41 | Kawakatsu Y, Nakayama H, Kaminoyama K, Igarashi K, Yasugi M, Kudoh H, Nagano AJ, Yano K, Kubo N, Kimura S (2017). A GLABRA1 ortholog on LG A9 controls trichome number in the Japanese leafy vegetables Mizuna and Mibuna(Brassica rapa L. subsp. nipposinica L. H. Bailey): evidence from QTL analysis. J Plant Res 130, 539-550. |
| 42 | Koch AJ, Meinhardt H (1994). Biological pattern formation: from basic mechanisms to complex structures.Rev Mod Phys 66, 1481-1507. |
| 43 | Kwak SH, Song SK, Lee MM, Schiefelbein J (2015). TORNADO1 regulates root epidermal patterning through the WEREWOLF pathway in Arabidopsis thaliana. Plant Signal Behav 10, e1103407. |
| 44 | Li Q, Cao CX, Zhang CJ, Zheng SS, Wang ZH, Wang LN, Ren ZH (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of tricho- mes uncovers a novel function for the homeodomain- leucine zipper I gene. J Exp Bot 66, 2515-2526. |
| 45 | Li WX, Chen TB, Chen Y, Lei M (2005). Role of trichome of Pteris vittata L. in arsenic hyperaccumulation. Sci China Ser C Life Sci 48, 148-154. |
| 46 | Li WQ, Wu JG, Weng SL, Zhang DP, Zhang YJ, Shi CH (2010). Characterization and fine mapping of the glabrous leaf and hull mutants (gl1) in rice(Oryza sativa L.). Plant Cell Rep 29, 617-627. |
| 47 | Li YQ, Shan XT, Gao RF, Yang S, Wang SC, Gao X, Wang L (2016). Two IIIf clade-bHLHs from freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci Rep 6, 30514. |
| 48 | Liu H, Zhou LH, Jiao JJ, Liu SB, Zhang ZM, Lu TJ, Xu F (2016a). Gradient mechanical properties facilitate Arabidopsis trichome as mechanosensor.ACS Appl Mater Interfaces 8, 9755-9761. |
| 49 | Liu M, Shi P, Fu XQ, Brodelius PE, Shen Q, Jiang WM, He Q, Tang KX (2016b). Characterization of a trichome- specific promoter of the aldehyde dehydrogenase 1 (ALDH1) gene in Artemisia annua. Plant Cell Tiss Organ Cult 126, 469-480. |
| 50 | Liu XW, Bartholomew E, Cai YL, Ren HZ (2016c). Tri- chome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L). Front Plant Sci 7, 1187. |
| 51 | Ma D, Hu Y, Yang CQ, Liu BL, Fang L, Wan Q, Liang WH, Mei GF, Wang LJ, Wang HP, Ding LY, Dong CG, Pan MQ, Chen JD, Wang S, Chen SQ, Cai CP, Zhu XF, Guan XY, Zhou BL, Zhu SJ, Wang JW, Guo WZ, Chen XY, Zhang TZ (2016). Genetic basis for glandular trichome formation in cotton.Nat Commun 7, 10456. |
| 52 | Maes L, Inzé D, Goossens A (2008). Functional specializa- tion of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves.Plant Physiol 148, 1453-1464. |
| 53 | Maes L, Van Nieuwerburgh FC, Zhang YS, Reed DW, Pollier J, Vande Casteele SR, Inzé D, Covello PS, Deforce DL, Goossens A (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol 189, 176-189. |
| 54 | Matías-Hernández L, Aguilar-Jaramillo AE, Cigliano RA, Sanseverino W, Pelaz S (2016a). Flowering and trichome development share hormonal and transcription factor regu- lation.J Exp Bot 67, 1209-1219. |
| 55 | Matías-Hernández L, Aguilar-Jaramillo AE, Osnato M, Weinstain R, Shani E, Suárez-López P, Pelaz S (2016b). TEMPRANILLO reveals the mesophyll as crucial for epidermal trichome formation.Plant Physiol 170, 1624-1639. |
| 56 | Matías-Hernández L, Jiang WM, Yang K, Tang KX, Brod- elius PE, Pelaz S (2017). AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome deve- lopment in Artemisia annua and Arabidopsis thaliana. Plant J 90, 520-534. |
| 57 | Mazie AR, Baum DA (2016). Clade-specific positive selection on a developmental gene:BRANCHLESS TRIC- HOME and the evolution of stellate trichomes in Physaria(Brassicaceae). Mol Phylogenet Evol 100, 31-40. |
| 58 | Mcdowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Des- cour A, Shi F, Larson M, Schilmiller A, An LL, Jones AD, Pichersky E, Soderlund CA, Gang DR (2011). Comparative functional genomic analysis of Solanum glan- dular trichome types. Plant Physiol 155, 524-539. |
| 59 | Meinhardt H, Gierer A (1974). Applications of a theory of biological pattern formation based on lateral inhibition.J Cell Sci 15, 321-346. |
| 60 | Mittal A, Balasubramanian R, Cao J, Singh P, Subrama nian S, Hicks G, Nothnagel EA, Abidi N, Janda J, Galbraith DW, Rock CD (2014). TOPOISOMERASE 6B is involved in chromatin remodelling associated with control of carbon partitioning into secondary metabolites and cell walls, and epidermal morphogenesis in Arabidopsis. J Exp Bot 65, 4217-4239. |
| 61 | Moro CF, Gaspar M, De Silva FR, Pattathil S, Hahn MG, Salgado I, Braga MR (2017). S-nitrosoglutathione promo- tes cell wall remodelling, alters the transcriptional profile and induces root hair formation in the hairless root hair defective 6 (rhd6) mutant of Arabidopsis thaliana. New Phytol 213, 1771-1786. |
| 62 | Ning PB, Wang JH, Zhou YL, Gao LF, Wang J, Gong CM (2016). Adaptional evolution of trichome in Caragana korshinskii to natural drought stress on the Loess Plateau, China. italic>Ecol Evol 6, 3786-3795. |
| 63 | Olfa B, Imen T, Mohamed C, Nasri-Ayachi MB (2016). Essential oil and trichome density from Origanum majorana L. shoots affected by leaf age and salinity. Biosci J 32, 238-245. |
| 64 | Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules.Cell 67, 483-493. |
| 65 | Patel JD, Wright RJ, Chandnani R, Goff VH, Ingles J, Paterson AH (2015). EMS-mutated cotton populations suggest overlapping genetic control of trichome and lint fiber variation.Euphytica 208, 597-608. |
| 66 | Pattanaik S, Patra B, Singh SK, Yuan L (2014). An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis.Front Plant Sci 5, 259. |
| 67 | Rakha M, Bouba N, Ramasamy S, Regnard JL, Hanson P (2017). Evaluation of wild tomato accessions (Solanum spp.) for resistance to two-spotted spider mite(Tetranychus urticae Koch) based on trichome type and acylsugar content. Genet Resour Crop Evol 64, 1011-1022. |
| 68 | Redonda-Martínez R, Villaseñor JL, Terrazas T (2016). Trichome diversity in the subtribe Leiboldiinae (Vernoni- eae, Asteraceae).J Torrey Bot Soc 143, 298-310. |
| 69 | Sato Y, Kudoh H (2017). Fine-scale frequency differentiation along a herbivory gradient in the trichome dimorphism of a wild Arabidopsis.Ecol Evol 7, 2133-2141. |
| 70 | Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis.EMBO J 21, 5036-5046. |
| 71 | Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL (2010). Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines.Plant J 62, 391-403. |
| 72 | Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC (2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol 191, 251-263. |
| 73 | Steede WT, Ma JM, Eickholt DP, Drake-Stowe KE, Kernodle SP, Shew D, Danehower DA, Lewis RS (2017). The tobacco trichome exudate Z-abienol and its relationship with plant resistance to Phytophthora nicotianae. Plant Dis 101, 1214-1221. |
| 74 | Stratmann JW, Bequette CJ (2016). Hairless but no longer clueless: understanding glandular trichome development.J Exp Bot 67, 5285-5287. |
| 75 | Tan JF, Walford SA, Dennis ES, Llewellyn D (2016). Trichomes control flower bud shape by linking together young petals.Nat Plants 2, 16093. |
| 76 | Tian DL, Peiffer M, De Moraes CM, Felton GW (2014). Roles of ethylene and jasmonic acid in systemic induced defense in tomato (Solanum lycopersicum) against Heli- coverpa zea. Planta 239, 577-589. |
| 77 | Tiwari P (2016). Recent advances and challenges in tri- chome research and essential oil biosynthesis in Mentha arvensis L. Ind Crops Prod 82, 141-148. |
| 78 | Tominaga R, Iwata M, Okada K, Wada T (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19, 2264-2277. |
| 79 | Tominaga-Wada R, Nukumizu Y, Sato S, Kato T, Tabata S, Wada T (2012). Functional divergence of MYB-related genes,WEREWOLF and AtMYB23 in Arabidopsis. Biosci Biotechnol Biochem 76, 883-887. |
| 80 | van Houten YM, Glas JJ, Hoogerbrugge H, Rothe J, Bolckmans KJF, Simoni S, van Arkel J, Alba JM, Kant MR, Sabelis MW (2013). Herbivory-associated degradation of tomato trichomes and its impact on biological control of Aculops lycopersici. Exp Appl Acarol 60, 127-138. |
| 81 | Vanhoutte B, Schenkels L, Ceusters J, De Proft MP (2017). Water and nutrient uptake in Vriesea cultivars: trichomes vs. roots. Environ Exp Bot 136, 21-30. |
| 82 | Vendemiatti E, Zsögön A, Silva GFFE, De Jesus FA, Cutri L, Figueiredo CRF, Tanaka FAO, Nogueira FTS, Peres LEP (2017). Loss of type-IV glandular trichomes is a hete- rochronic trait in tomato and can be reverted by promoting juvenility.Plant Sci 259, 35-47. |
| 83 | Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM (2009). The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize.Plant J 59, 883-894. |
| 84 | Vitarelli NC, Riina R, Cassino MF, Meira RMSA (2016). Trichome-like emergences in Croton of Brazilian highland rock outcrops: evidences for atmospheric water uptake.Perspect Plant Ecol Evol Syst 22, 23-35. |
| 85 | Wada T, Tominaga-Wada R (2015). CAPRICE family genes control flowering time through both promoting and rep- ressing CONSTANS and FLOWERING LOCUS T expre- ssion. Plant Sci 241, 260-265. |
| 86 | Wang G, Feng HJ, Sun JL, Du XM (2013). Induction of cotton ovule culture fibre branching by co-expression of cotton BTL, cotton SIM, and Arabidopsis STI genes. J Exp Bot 64, 4157-4168. |
| 87 | Wang Q, Reddy VA, Panicker D, Mao HZ, Kumar N, Rajan C, Venkatesh PN, Chua NH, Sarojam R (2016a). Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint (Mentha spicata). Plant Biotechnol J 14, 1619-1632. |
| 88 | Wang YL, Nie JT, Chen HM, Guo CL, Pan J, He HL, Pan JS, Cai R (2016b). Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor Appl Genet 129, 305-316. |
| 89 | Weinhold A, Baldwin IT (2011). Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc Natl Acad Sci USA 108, 7855-7859. |
| 90 | Xiao L, Tan HX, Zhang L (2016). Artemisia annua glandular secretory trichomes: the biofactory of antimalarial agent artemisinin. Sci Bull 61, 26-36. |
| 91 | Xie Y, Tan HJ, Ma ZX, Huang JR (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL- 2 and JAZ suppressors of the MYB/bHLH/WD40 complex inArabidopsis thaliana. Mol Plant 9, 711-721. |
| 92 | Yan TX, Chen MH, Shen Q, Li L, Fu XQ, Pan QF, Tang YL, Shi P, Lv ZY, Jiang WM, Ma YN, Hao XL, Sun XF, Tang KX (2017). HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol 213, 1145-1155. |
| 93 | Yanagisawa M, Desyatova AS, Belteton SA, Mallery EL, Turner JA, Szymanski DB (2015). Patterning mechanisms of cytoskeletal and cell wall systems during leaf trichome morphogenesis.Nat Plants 1, 15014. |
| 94 | Yu N, Cai WJ, Wang SC, Shan CM, Wang LJ, Chen XY (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322-2335. |
| 95 | Zhang CH, Mallery EL, Schlueter J, Huang SJ, Fan YR, Brankle S, Staiger CJ, Szymanski DB (2008). Arabidop- sis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20, 995-1011. |
| 96 | Zhao Q, Chen XY (2016). Development: a new function of plant trichomes.Nat Plants 2, 16096. |
| 97 | Zheng KJ, Tian HN, Hu QN, Guo HY, Yang L, Cai L, Wang XT, Liu B, Wang SC (2016). Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci Rep 6, 19254. |
| 98 | Zhou LH, Liu SB, Wang PF, Lu TJ, Xu F, Genin GM, Pickard BG (2017). The Arabidopsis trichome is an active mechanosensory switch.Plant Cell Environ 40, 611-621. |
| 99 | Zhou WQ, Wang YC, Wu ZL, Luo L, Liu P, Yan LF, Hou SW (2016). Homologs of SCAR/WAVE complex compon- ents are required for epidermal cell morphogenesis in rice.J Exp Bot 67, 4311-4323. |
| 100 | Zhou ZJ, An LJ, Sun LL, Gan YB (2012). ZFP5 encodes a functionally equivalent GIS protein to control trichome ini- tiation. Plant Signal Behav 7, 28-30. |
| 101 | Zou JJ, Zheng ZY, Xue S, Li HH, Wang YR, Le J (2016). The role of Arabidopsis Actin-Related Protein 3 in amyloplast sedimentation and polar auxin transport in root gravitropism.J Exp Bot 67, 5325-5337. |
| [1] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 崔娟, 于晓玉, 于跃娇, 梁铖玮, 孙健, 陈温福. 影响中国东北和日本粳稻食味品质差异的质构因素及其遗传基础解析[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 王子韵, 吕燕文, 肖钰, 吴超, 胡新生. 植物基因表达调控与进化机制研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [6] | 陆珍, 谢光杰, Qaisar KHAN, 覃英, 黄毓燕, 郭道君, 杨婷婷, 杨丽涛, 邢永秀, 李杨瑞, 王震. 伯克霍尔德菌通过改善生理适应性及调节铝响应基因的表达增强甘蔗对铝胁迫的耐受性[J]. 植物生态学报, 2025, 49(3): 475-487. |
| [7] | 张如礼, 李德铢, 张玉霄. 短穗竹居群遗传结构及气候适应性分析[J]. 植物学报, 2025, 60(3): 407-424. |
| [8] | 仝淼, 王欢, 张文双, 王超, 宋建潇. 重金属污染土壤中细菌抗生素抗性基因分布特征[J]. 生物多样性, 2025, 33(3): 24101-. |
| [9] | 杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析[J]. 植物学报, 2025, 60(3): 377-392. |
| [10] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [11] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [12] | 曹东, 李焕龙, 彭扬, 魏存争. 植物基因组大小与性状关系的研究进展[J]. 生物多样性, 2025, 33(2): 24192-. |
| [13] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [14] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [15] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||