

植物学报 ›› 2023, Vol. 58 ›› Issue (1): 77-89.DOI: 10.11983/CBB22138 cstr: 32102.14.CBB22138
所属专题: 杂粮生物学专辑 (2023年58卷1期)
李晓明1, 王兰芬1, 唐永生2, 常玉洁1, 张菊香2, 王述民1, 武晶1,*( )
)
收稿日期:2022-06-30
接受日期:2022-09-19
出版日期:2023-01-01
发布日期:2023-01-05
通讯作者:
*E-mail: wujing@caas.cn
作者简介:†共同第一作者
基金资助:
Xiaoming Li1, Lanfen Wang1, Yongsheng Tang2, Yujie Chang1, Juxiang Zhang2, Shumin Wang1, Jing Wu1,*( )
)
Received:2022-06-30
Accepted:2022-09-19
Online:2023-01-01
Published:2023-01-05
Contact:
*E-mail: wujing@caas.cn
About author:†These authors contributed equally to this paper
摘要: 普通菜豆(Phaseolus vulgaris)具有丰富的营养价值, 菜豆象(Acanthoscelides obtectus)是危害菜豆的主要害虫, 利用抗虫种质资源防治菜豆象是最安全且经济有效的方法。该研究利用改良的室内人工接虫方法, 对625份普通菜豆种质资源进行2次菜豆象抗性重复鉴定, 筛选出2份抗性稳定且种子受害率均在10%以下的高抗种质。利用种子受害率和蛀孔总数的表型数据, 基于3 767 432个SNP标记进行全基因组关联分析, 鉴定出15个与种子受害率相关的显著关联遗传位点, 8个与蛀孔总数相关的显著关联位点, 解释了4.54%-5.56%的表型变异。在候选位点筛选出包括编码蛋白酶抑制剂、凝集素和过氧化物酶等在内的20个与抗虫防御相关的候选基因。
李晓明, 王兰芬, 唐永生, 常玉洁, 张菊香, 王述民, 武晶. 普通菜豆抗菜豆象性状的全基因组关联分析. 植物学报, 2023, 58(1): 77-89.
Xiaoming Li, Lanfen Wang, Yongsheng Tang, Yujie Chang, Juxiang Zhang, Shumin Wang, Jing Wu. Genome-wide Association Analysis of Resistance to Acanthoscelides obtectus in Common Bean. Chinese Bulletin of Botany, 2023, 58(1): 77-89.
| Gene pool | Outside of Chinaa | Inside of Chinab | All | ||
|---|---|---|---|---|---|
| BL | L | BL | L | ||
| Andean | 33 | 16 | 27 | 206 | 282 |
| Mesoamerican | 64 | 11 | 20 | 248 | 343 |
| All | 97 | 27 | 47 | 454 | 625 |
表1 625份普通菜豆种质资源的来源及分类
Table 1 Origin and classification of 625 common bean germplasm resources
| Gene pool | Outside of Chinaa | Inside of Chinab | All | ||
|---|---|---|---|---|---|
| BL | L | BL | L | ||
| Andean | 33 | 16 | 27 | 206 | 282 |
| Mesoamerican | 64 | 11 | 20 | 248 | 343 |
| All | 97 | 27 | 47 | 454 | 625 |
| Rating scale | PDS (%) | Resistant evaluation |
|---|---|---|
| 1 | ≤10 | Highly resistance (HR) |
| 3 | 10<PDS≤35 | Resistance (R) |
| 5 | 35<PDS≤65 | Medium resistance (MR) |
| 7 | 65<PDS≤90 | Susceptible (S) |
| 9 | >90 | Highly susceptible (HS) |
表2 普通菜豆种质资源抗菜豆象的分级评价标准
Table 2 Evaluation standard of resistance grade of common bean germplasm resources
| Rating scale | PDS (%) | Resistant evaluation |
|---|---|---|
| 1 | ≤10 | Highly resistance (HR) |
| 3 | 10<PDS≤35 | Resistance (R) |
| 5 | 35<PDS≤65 | Medium resistance (MR) |
| 7 | 65<PDS≤90 | Susceptible (S) |
| 9 | >90 | Highly susceptible (HS) |
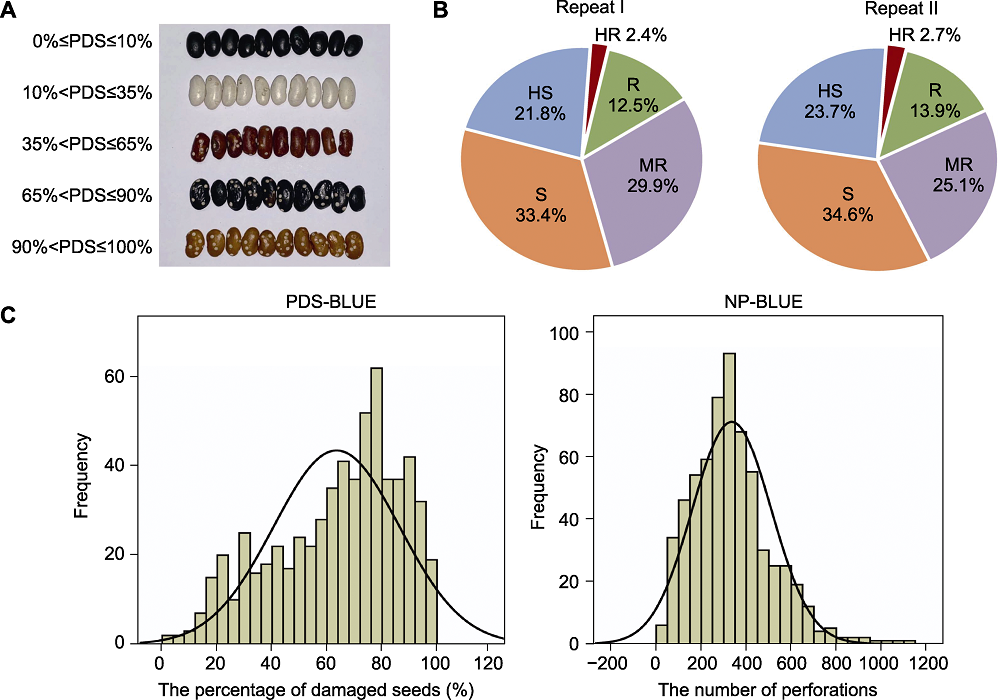
图1 普通菜豆种质资源菜豆象抗性2次重复鉴定结果 (A) 不同抗性级别菜豆的受侵染情况; (B) 2次重复鉴定的菜豆象抗性鉴定结果; (C) 625份普通菜豆种质2次重复鉴定的种子受害率和蛀孔总数最佳线性无偏估计值(BLUE值)的分布直方图。Repeat I: 第1次重复鉴定; Repeat II: 第2次重复鉴定; PDS: 种子受害率; NP: 蛀孔总数
Figure 1 Results of two repeated identification of the resistance of common bean germplasm resources to Acanthoscelides obtectus (A) Infestation by A. obtectus of common beans with different resistance levels; (B) Results of bruchid resistance in two repeated identifications; (C) Distribution histogram of the best linear unbiased estimators (BLUE) values of PDS and NP in 625 common beans of two repeated identifications. Repeat I: The first repeated identification; Repeat II: The second repeated identification; PDS: The percentage of damaged seeds; NP: The number of perforations
| Traits | Min | Median | Max | Mean | SD (%) | Kurtosis | Skewness | CV (%) |
|---|---|---|---|---|---|---|---|---|
| PDS I (%) | 1.00 | 67.50 | 100 | 64.14 | 25.65 | -0.71 | -0.45 | 40.00 |
| NP I | 3.00 | 263.50 | 1224 | 294.12 | 181.20 | 1.48 | 0.96 | 61.61 |
| PDS II (%) | 0.00 | 72.00 | 100 | 66.44 | 27.17 | -0.76 | -0.62 | 40.89 |
| NP II | 0.00 | 329.00 | 1816 | 376.20 | 254.91 | 3.12 | 1.37 | 67.76 |
| PDS-BLUE (%) | 4.00 | 70.00 | 100 | 65.44 | 0.23 | -0.63 | -0.58 | 35.07 |
| NP-BLUE | 0.56 | 318.00 | 1126 | 335.55 | 174.94 | 1.56 | 0.92 | 52.02 |
表3 625份普通菜豆种子受害率和蛀孔总数的表型特征
Table 3 Phenotypic characteristics of PDS and NP in 625 common beans
| Traits | Min | Median | Max | Mean | SD (%) | Kurtosis | Skewness | CV (%) |
|---|---|---|---|---|---|---|---|---|
| PDS I (%) | 1.00 | 67.50 | 100 | 64.14 | 25.65 | -0.71 | -0.45 | 40.00 |
| NP I | 3.00 | 263.50 | 1224 | 294.12 | 181.20 | 1.48 | 0.96 | 61.61 |
| PDS II (%) | 0.00 | 72.00 | 100 | 66.44 | 27.17 | -0.76 | -0.62 | 40.89 |
| NP II | 0.00 | 329.00 | 1816 | 376.20 | 254.91 | 3.12 | 1.37 | 67.76 |
| PDS-BLUE (%) | 4.00 | 70.00 | 100 | 65.44 | 0.23 | -0.63 | -0.58 | 35.07 |
| NP-BLUE | 0.56 | 318.00 | 1126 | 335.55 | 174.94 | 1.56 | 0.92 | 52.02 |
| Accession No. | Origin | 100 seeds weight (g) | Repeat I | Repeat II | ||
|---|---|---|---|---|---|---|
| PDS (%) | NP | PDS (%) | NP | |||
| Longyundou7 | China-Heilongjiang | 17.29 | 4 | 21 | 4 | 11 |
| I2K03033 | USA | 17.34 | 10 | 30 | 6 | 24 |
表4 2次重复鉴定的2份稳定高抗的普通菜豆种质
Table 4 Two germplasms of common beans with stable high resistance in two repeated identifications
| Accession No. | Origin | 100 seeds weight (g) | Repeat I | Repeat II | ||
|---|---|---|---|---|---|---|
| PDS (%) | NP | PDS (%) | NP | |||
| Longyundou7 | China-Heilongjiang | 17.29 | 4 | 21 | 4 | 11 |
| I2K03033 | USA | 17.34 | 10 | 30 | 6 | 24 |
| Traits | Min | Median | Max | Mean | SD (%) | Kurtosis | Skewness | CV (%) |
|---|---|---|---|---|---|---|---|---|
| SW I (g) | 10.80 | 34.21 | 80.40 | 34.95 | 12.93 | -0.57 | 0.36 | 37.00 |
| SW II (g) | 7.08 | 31.62 | 70.23 | 33.18 | 12.35 | -0.44 | 0.42 | 37.22 |
| SL I (mm) | 6.30 | 12.20 | 17.80 | 12.30 | 2.51 | -1.01 | 0.05 | 20.40 |
| SL II (mm) | 6.25 | 13.89 | 15.25 | 14.94 | 3.02 | -0.93 | -0.06 | 20.19 |
| SWI I (mm) | 4.20 | 7.10 | 15.60 | 7.11 | 1.13 | 9.79 | 1.59 | 15.91 |
| SWI II (mm) | 4.88 | 8.70 | 13.50 | 8.66 | 1.40 | -0.25 | 0.19 | 16.20 |
| SH I (mm) | 3.80 | 5.50 | 8.50 | 5.62 | 0.85 | 0.71 | 0.91 | 15.03 |
| SH II (mm) | 3.70 | 5.40 | 8.80 | 5.49 | 0.82 | 0.58 | 0.74 | 14.90 |
表5 625份普通菜豆籽粒大小的性状特征
Table 5 Characteristics of seeds size in 625 common beans
| Traits | Min | Median | Max | Mean | SD (%) | Kurtosis | Skewness | CV (%) |
|---|---|---|---|---|---|---|---|---|
| SW I (g) | 10.80 | 34.21 | 80.40 | 34.95 | 12.93 | -0.57 | 0.36 | 37.00 |
| SW II (g) | 7.08 | 31.62 | 70.23 | 33.18 | 12.35 | -0.44 | 0.42 | 37.22 |
| SL I (mm) | 6.30 | 12.20 | 17.80 | 12.30 | 2.51 | -1.01 | 0.05 | 20.40 |
| SL II (mm) | 6.25 | 13.89 | 15.25 | 14.94 | 3.02 | -0.93 | -0.06 | 20.19 |
| SWI I (mm) | 4.20 | 7.10 | 15.60 | 7.11 | 1.13 | 9.79 | 1.59 | 15.91 |
| SWI II (mm) | 4.88 | 8.70 | 13.50 | 8.66 | 1.40 | -0.25 | 0.19 | 16.20 |
| SH I (mm) | 3.80 | 5.50 | 8.50 | 5.62 | 0.85 | 0.71 | 0.91 | 15.03 |
| SH II (mm) | 3.70 | 5.40 | 8.80 | 5.49 | 0.82 | 0.58 | 0.74 | 14.90 |
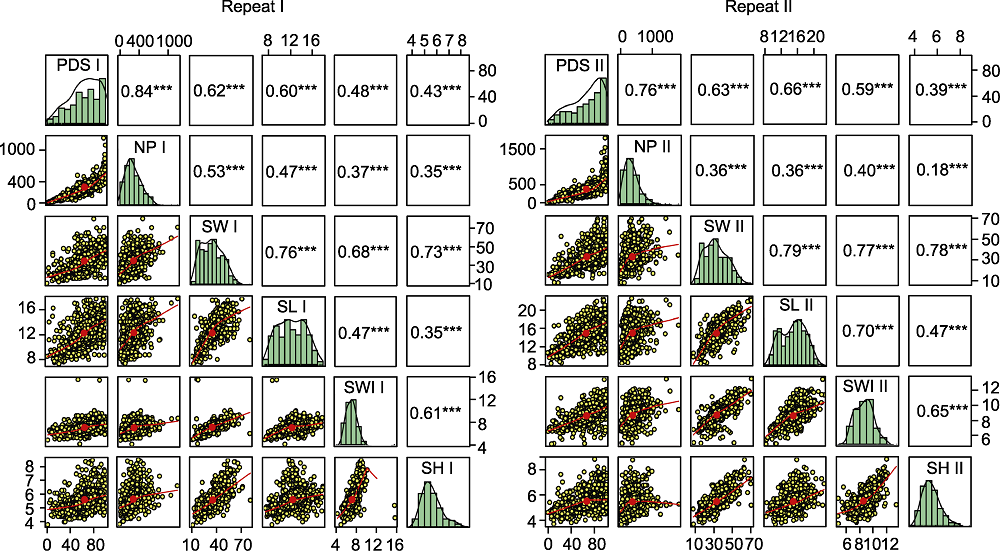
图2 2次重复鉴定的种质资源中菜豆籽粒大小与菜豆象抗性的相关性分析 PDS I、NP I、PDS II和NP II同表3。SW I、SW II、SL I、SL II、SWI I、SWI II、SH I和SH II同表5。*** P<0.001
Figure 2 Correlation analysis for seeds size and bruchid resistance of germplasm resources in two repeated identifications PDS I, NP I, PDS II and NP II see Table 3. SW I, SW II, SL I, SL II, SWI I, SWI II, SH I and SH II see Table 5. *** P<0.001

图4 PDS和NP全基因组关联分析的曼哈顿图 (A) 混合线性模型(MLM)下种子受害率(PDS-BLUE)的曼哈顿图(左)和Q-Q图(右); (B) 混合线性模型下蛀孔总数(NP-BLUE)的曼哈顿图(左)和Q-Q图(右)。横坐标为染色体, 纵坐标为LOD效应值。黑色虚线为阈值(-Log10 p=6), 超过虚线的点为显著关联位点, 用红色标注。PDS和NP同图1。
Figure 4 Manhattan plot of PDS and NP by genome-wide association study (A) Manhattan plot (left) and Q-Q plot (right) of PDS-BLUE by mixed linear model (MLM); (B) Manhattan plot (left) and Q-Q plot (right) of NP-BLUE by the MLM. The abscissa is the chromosome, and the ordinate is the LOD effect value. The black dotted line is the threshold value (-Log10 p=6), and the points exceeding the threshold are the significant association sites, highlighting in red. PDS and NP see Figure 1.
| Traits | Locus | Peak SNP marker | Chromosome | Position (bp) | p-value | R2 (%) |
|---|---|---|---|---|---|---|
| PDS | Loc_1 | Pv01_43986475 | Pv01 | 43986475 | 1E-07 | 5.24 |
| Loc_2 | Pv01_49901915 | Pv01 | 49901915 | 8.15E-07 | 4.54 | |
| Loc_3 | Pv03_1235524 | Pv03 | 1235524 | 2.25E-07 | 4.97 | |
| Loc_4 | Pv03_6664019 | Pv03 | 6664019 | 4.46E-07 | 4.74 | |
| Loc_5 | Pv03_11569042 | Pv03 | 11569042 | 2.19E-07 | 4.98 | |
| Loc_6 | Pv03_37792879 | Pv03 | 37792879 | 5.51E-07 | 4.67 | |
| Loc_7 | Pv05_522813 | Pv05 | 522813 | 3.89E-07 | 4.79 | |
| Loc_8 | Pv05_5075510 | Pv05 | 5075510 | 3.54E-07 | 4.82 | |
| Loc_9 | Pv05_22022919 | Pv05 | 22022919 | 3.89E-08 | 5.56 | |
| Loc_10 | Pv06_19260606 | Pv06 | 19260606 | 7.22E-07 | 4.58 | |
| Loc_11 | Pv07_386049 | Pv07 | 386049 | 6.01E-07 | 4.64 | |
| Loc_12 | Pv08_13992928 | Pv08 | 13992928 | 6.42E-07 | 4.62 | |
| Loc_13 | Pv08_43139505 | Pv08 | 43139505 | 5.06E-07 | 4.70 | |
| Loc_14 | Pv10_9574825 | Pv10 | 9574825 | 2.3E-07 | 4.96 | |
| Loc_15 | Pv11_11777960 | Pv11 | 11777960 | 2.62E-07 | 4.92 | |
| NP | Loc_16 | Pv02_36436725 | Pv02 | 36436725 | 5.26E-07 | 4.74 |
| Loc_17 | Pv03_23935240 | Pv03 | 23935240 | 5.31E-07 | 4.74 | |
| Loc_18 | Pv04_9609212 | Pv04 | 9609212 | 2.01E-07 | 5.07 | |
| Loc_19 | Pv04_10133957 | Pv04 | 10133957 | 5.13E-07 | 4.75 | |
| Loc_20 | Pv04_44834996 | Pv04 | 44834996 | 7.56E-07 | 4.62 | |
| Loc_21 | Pv10_8170537 | Pv10 | 8170537 | 6.45E-07 | 4.68 | |
| Loc_22 | Pv11_13013375 | Pv11 | 13013375 | 2.37E-07 | 5.01 | |
| Loc_23 | Pv11_19511385 | Pv11 | 19511385 | 7.45E-07 | 4.63 |
表6 全基因组关联分析(GWAS)鉴定的菜豆象抗性相关位点
Table 6 All loci related with bruchid resistance detected by genome-wide association study (GWAS)
| Traits | Locus | Peak SNP marker | Chromosome | Position (bp) | p-value | R2 (%) |
|---|---|---|---|---|---|---|
| PDS | Loc_1 | Pv01_43986475 | Pv01 | 43986475 | 1E-07 | 5.24 |
| Loc_2 | Pv01_49901915 | Pv01 | 49901915 | 8.15E-07 | 4.54 | |
| Loc_3 | Pv03_1235524 | Pv03 | 1235524 | 2.25E-07 | 4.97 | |
| Loc_4 | Pv03_6664019 | Pv03 | 6664019 | 4.46E-07 | 4.74 | |
| Loc_5 | Pv03_11569042 | Pv03 | 11569042 | 2.19E-07 | 4.98 | |
| Loc_6 | Pv03_37792879 | Pv03 | 37792879 | 5.51E-07 | 4.67 | |
| Loc_7 | Pv05_522813 | Pv05 | 522813 | 3.89E-07 | 4.79 | |
| Loc_8 | Pv05_5075510 | Pv05 | 5075510 | 3.54E-07 | 4.82 | |
| Loc_9 | Pv05_22022919 | Pv05 | 22022919 | 3.89E-08 | 5.56 | |
| Loc_10 | Pv06_19260606 | Pv06 | 19260606 | 7.22E-07 | 4.58 | |
| Loc_11 | Pv07_386049 | Pv07 | 386049 | 6.01E-07 | 4.64 | |
| Loc_12 | Pv08_13992928 | Pv08 | 13992928 | 6.42E-07 | 4.62 | |
| Loc_13 | Pv08_43139505 | Pv08 | 43139505 | 5.06E-07 | 4.70 | |
| Loc_14 | Pv10_9574825 | Pv10 | 9574825 | 2.3E-07 | 4.96 | |
| Loc_15 | Pv11_11777960 | Pv11 | 11777960 | 2.62E-07 | 4.92 | |
| NP | Loc_16 | Pv02_36436725 | Pv02 | 36436725 | 5.26E-07 | 4.74 |
| Loc_17 | Pv03_23935240 | Pv03 | 23935240 | 5.31E-07 | 4.74 | |
| Loc_18 | Pv04_9609212 | Pv04 | 9609212 | 2.01E-07 | 5.07 | |
| Loc_19 | Pv04_10133957 | Pv04 | 10133957 | 5.13E-07 | 4.75 | |
| Loc_20 | Pv04_44834996 | Pv04 | 44834996 | 7.56E-07 | 4.62 | |
| Loc_21 | Pv10_8170537 | Pv10 | 8170537 | 6.45E-07 | 4.68 | |
| Loc_22 | Pv11_13013375 | Pv11 | 13013375 | 2.37E-07 | 5.01 | |
| Loc_23 | Pv11_19511385 | Pv11 | 19511385 | 7.45E-07 | 4.63 |
| Traits | Gene | Chromosome | Locus | Gene annotation or coding protein |
|---|---|---|---|---|
| PDS | Phvul.001G239200 | Pv01 | Loc_2 | Concanavalin A-like lectin family protein |
| Phvul.003G014200 | Pv03 | Loc_3 | Hydroxyproline-rich glycoprotein family protein | |
| Phvul.003G169100 | Pv03 | Loc_6 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | |
| Phvul.005G005800 | Pv05 | Loc_7 | WRKY DNA-binding protein 2 | |
| Phvul.005G046000 | Pv05 | Loc_8 | Peroxidase superfamily protein | |
| Phvul.006G073200 | Pv06 | Loc_10 | Hydroxyproline-rich glycoprotein family protein | |
| Phvul.008G113100 | Pv08 | Loc_12 | Kunitz family trypsin and protease inhibitor protein | |
| Phvul.008G113900 | Pv08 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | ||
| Phvul.010G057000 | Pv10 | Loc_14 | S-locus lectin protein kinase family protein | |
| Phvul.010G057100 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057300 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057500 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057900 | Pv10 | WRKY DNA-binding protein 70 | ||
| NP | Phvul.002G205700 | Pv02 | Loc_16 | Defensin-like (DEFL) family protein |
| Phvul.002G205900 | Pv02 | Defensin-like (DEFL) family protein | ||
| Phvul.003G099300 | Pv03 | Loc_17 | S-locus lectin protein kinase family protein | |
| Phvul.004G067300 | Pv04 | Loc_18 | Sterol | |
| Phvul.004G068800 | Pv04 | Loc_19 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | |
| Phvul.011G108700 | Pv11 | Loc_22 | S-locus lectin protein kinase family protein | |
| Phvul.011G119200 | Pv11 | Loc_23 | Concanavalin A-like lectin protein kinase family protein |
表7 筛选获得的候选基因信息
Table 7 Information of candidate genes by screening
| Traits | Gene | Chromosome | Locus | Gene annotation or coding protein |
|---|---|---|---|---|
| PDS | Phvul.001G239200 | Pv01 | Loc_2 | Concanavalin A-like lectin family protein |
| Phvul.003G014200 | Pv03 | Loc_3 | Hydroxyproline-rich glycoprotein family protein | |
| Phvul.003G169100 | Pv03 | Loc_6 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | |
| Phvul.005G005800 | Pv05 | Loc_7 | WRKY DNA-binding protein 2 | |
| Phvul.005G046000 | Pv05 | Loc_8 | Peroxidase superfamily protein | |
| Phvul.006G073200 | Pv06 | Loc_10 | Hydroxyproline-rich glycoprotein family protein | |
| Phvul.008G113100 | Pv08 | Loc_12 | Kunitz family trypsin and protease inhibitor protein | |
| Phvul.008G113900 | Pv08 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | ||
| Phvul.010G057000 | Pv10 | Loc_14 | S-locus lectin protein kinase family protein | |
| Phvul.010G057100 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057300 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057500 | Pv10 | S-locus lectin protein kinase family protein | ||
| Phvul.010G057900 | Pv10 | WRKY DNA-binding protein 70 | ||
| NP | Phvul.002G205700 | Pv02 | Loc_16 | Defensin-like (DEFL) family protein |
| Phvul.002G205900 | Pv02 | Defensin-like (DEFL) family protein | ||
| Phvul.003G099300 | Pv03 | Loc_17 | S-locus lectin protein kinase family protein | |
| Phvul.004G067300 | Pv04 | Loc_18 | Sterol | |
| Phvul.004G068800 | Pv04 | Loc_19 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | |
| Phvul.011G108700 | Pv11 | Loc_22 | S-locus lectin protein kinase family protein | |
| Phvul.011G119200 | Pv11 | Loc_23 | Concanavalin A-like lectin protein kinase family protein |
| [1] | 江兆春, 张忠民, 耿坤, 余杰颖 (2018). 菜豆象入侵我国的风险性评价. 贵州农业科学 46, 47-50. |
| [2] | 阮期平, 周立, 郑远旗 (2000). PGIP在植物抗病方面的研究进展. 植物学通报 17, 60-63. |
| [3] | 申智慧, 刘春, 杨洪, 郑松, 罗全丽, 徐本刚 (2014). 菜豆象发生规律与防治措施. 耕作与栽培 3, 47-48. |
| [4] | 宋凤鸣, 郑重, 葛起新 (1992). 富含羟脯氨酸糖蛋白在植物-病原物相互作用中的积累、作用及调控. 植物生理学通讯 28, 141-145. |
| [5] | 孙海燕, 罗兵, 喻德跃 (2005). 次生代谢抗虫基因工程研究进展. 安徽农业科学 33, 1906-1907. |
| [6] | 王辉, 李琳, 梁正, 吴一江 (2013). 菜豆象检疫及防治研究进展. 农业灾害研究 3(9), 8-12. |
| [7] | 吴长松, 冯明义, 李海, 姜府文, 赵芳, 龙贵兴, 丁昭兵, 卢云昌, 彭瑶, 廖华刚, 张国升, 颜兴, 陶惠, 文静, 高强 (2019). 毕节市菜豆象疫情发生危害及储存期防控措施初探. 植物检疫 33, 59-61. |
| [8] | 杨璐, 戴仁怀, 杨洪, 江兆春, 李敏, 杨燕琼 (2022). 甲酸乙酯和异硫氰酸甲酯混用对菜豆象的熏蒸活性研究. 山地农业生物学报 41, 79-83. |
| [9] | 杨乃博, 伍苏然, 沈林波, 张树珍, 杨本鹏 (2014). 植物抗虫性研究概况. 热带农业科学 34(9), 61-68, 89. |
| [10] | 周晓宇, 陈杰, 杨敬, 黄海涛, 宋翔 (2010). 植物凝集素及其在抗虫基因工程中的应用. 山地农业生物学报 29, 255-260. |
| [11] | Apostolova ED, Palagacheva NG, Svetleva DL, Mateeva AV (2013). Investigations on the resistance of some Bulgarian common bean genotypes towards bean weevil (Acanthoscelides obtectus Say). J Cent Eur Agric 14, 1547-1557. |
| [12] | Azizoglu U (2018). Biochemical properties of Turkish common beans and their resistance against bean weevil Acanthoscelides obtectus (Coleoptera: Bruchidae). Ar-thropod-plant Inte 12, 283-290. |
| [13] |
Blair MW, Muñoz C, Buendía HF, Flower J, Bueno JM, Cardona C (2010a). Genetic mapping of microsatellite markers around the Arcelin bruchid resistance locus in common bean. Theor Appl Genet 121, 393-402.
DOI URL |
| [14] |
Blair MW, Prieto S, Díaz LM, Buendía HF, Cardona C (2010b). Linkage disequilibrium at the APA insecticidal seed protein locus of common bean (Phaseolus vulgaris L.). BMC Plant Biol 10, 79.
DOI URL |
| [15] |
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633-2635.
DOI PMID |
| [16] |
Buchler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li HH, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Villeda HS, da Silva HS, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu JM, Zhang ZW, Kresovich S, Mcmullen MD (2009). The genetic architecture of maize flowering time. Science 325, 714-718.
DOI PMID |
| [17] |
Chen KC, Lin CY, Kuan CC, Sung HY, Chen CS (2002). A novel defensin encoded by a mungbean cDNA exhibits insecticidal activity against bruchid. J Agric Food Chem 50, 7258-7263.
DOI URL |
| [18] |
Cormier F, Le Gouis J, Dubreuil P, Lafarge S, Praud S (2014). A genome-wide identification of chromosomal re-gions determining nitrogen use efficiency components in wheat (Triticum aestivum L.). Theor Appl Genet 127, 2679-2693.
DOI PMID |
| [19] |
De Andrade EKV, Rodrigues R, da Costa Vieira Bard G, da Silva Pereira L, Baptista KEV, Cavalcanti TFM, Oliveira AEA, Souza TAM, Gomes VM (2020). Identifi-cation, biochemical characterization and biological role of defense proteins from common bean genotypes seeds in response to Callosobruchus maculatus infestation. J Stored Prod Res 87, 101580.
DOI URL |
| [20] |
Duan CX, Zhu ZD, Li WC, Bao SY, Wang XM (2017). Ge-netic diversity and differentiation of Acanthoscelides ob-tectus Say (Coleoptera: Bruchidae) populations in China. Agric For Entomol 19, 113-121.
DOI URL |
| [21] | Duarte MAG, Cabral GB, Ibrahim AB, Aragão FJL (2018). An overview of the APA locus and Arcelin proteins and their biotechnological potential in the control of bruchids. Agri Gene 8, 57-62. |
| [22] |
Gautam AK, Shrivastava N, Sharma B, Bhagyawant SS (2018). Current scenario of legume lectins and their prac-tical applications. J Crop Sci Biotechnol 21, 217-227.
DOI |
| [23] |
Goossens A, Quintero C, Dillen W, DeRycke R, Valor JF, De Clercq J, Van Montagu M, Cardona C, Angenon G (2000). Analysis of bruchid resistance in the wild common bean accession G02771: no evidence for insecticidal ac-tivity of Arcelin 5. J Exp Bot 51, 1229-1236.
PMID |
| [24] |
Gupta M, Sharma P, Nath AK (2014). Purification of a novel α-amylase inhibitor from local Himalayan bean (Phaseo-lus vulgaris) seeds with activity towards bruchid pests and human salivary amylase. J Food Sci Technol 51, 1286-1293.
DOI URL |
| [25] | Hategekimana A, Erler F (2020). Fecundity and fertility inhibition effects of some plant essential oils and their major components against Acanthoscelides obtectus Say (Coleoptera: Bruchidae). J Plant Dis Protect 127, 615-623. |
| [26] |
Huang XH, Zhao Y, Wei XH, Li CY, Wang AH, Zhao Q, Li WJ, Guo YL, Deng LW, Zhu CR, Fan DL, Lu YQ, Weng QJ, Liu KY, Zhou TY, Jing YF, Si LZ, Dong GJ, Huang T, Lu TT, Feng Q, Qian Q, Li JY, Han B (2012). Geno-me-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44, 32-39.
DOI |
| [27] |
Iturralde-García RD, Castañé C, Wong-corral FJ, Riu-davets J (2020). Biological control of Acanthoscelides obtectus and Zabrotes subfasciatus in stored dried beans. BioControl 65, 693-701.
DOI |
| [28] |
Jia MJ, Yang LJ, Zhang W, Rosewarne G, Li JH, Yang EN, Chen L, Wang WX, Liu YK, Tong HW, He WJ, Zhang YQ, Zhu ZW, Gao CB (2020). Genome-wide as-sociation analysis of stripe rust resistance in modern Chinese wheat. BMC Plant Biol 20, 491.
DOI |
| [29] |
Jiménez JC, de la Fuente M, Ordás B, Domínguez LEG, Malvae RA (2017). Resistance categories to Acanthosce-lides obtectus (Coleoptera: Bruchidae) in tepary bean (Phaseolus acutifolius), new sources of resistance for dry bean (Phaseolus vulgaris) breeding. Crop Prot 98, 255-266.
DOI URL |
| [30] | Joshi J, Pandurangan S, Diapari M, Marsolais F (2017). Comparison of gene families:seed storage and other seed proteins. In: de la Vega MP, Santalla M, Marsolais F, eds. The Common Bean Genome. Cham: Springer. pp. 201-217. |
| [31] |
Kamfwa K, Beaver JS, Cichy KA, Kelly JD (2018). QTL mapping of resistance to bean weevil in common bean. Crop Sci 58, 2370-2378.
DOI URL |
| [32] |
Kornegay JL, Cardona C (1991). Inheritance of resistance to Acanthoscelides obtectus in a wild common bean ac-cession crossed to commercial bean cultivars. Euphytica 52, 103-111.
DOI |
| [33] |
Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, Balint-Kurti PJ, Holland JB (2011). Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested as-sociation mapping population. Nat Genet 43, 163-168.
DOI |
| [34] | Kusolwa PM, Myers JR (2011). Seed storage proteins ARL2 and its variants from the apalocus of wild tepary bean G40199 confers resistance to Acanthoscelides ob-tectus when expressed in common beans. Afr Crop Sci J 19, 255-265. |
| [35] |
Kusolwa PM, Myers JR, Porch TG, Trukhina Y, González-Vélez A, Beaver JS (2016). Registration of AO- 1012-29-3-3A red kidney bean germplasm line with bean weevil, BCMV, and BCMNV resistance. J Plant Regist 10, 149-153.
DOI URL |
| [36] | Li B, Chen SL, Qin C, Xiao J (2016). Research progress on germplasm resources of common bean. Agric Sci Technol 17, 2572-2577. |
| [37] |
Li XM, Tang YS, Wang LF, Chang YJ, Wu J, Wang SM (2022). QTL mapping and identification of genes associated with the resistance to Acanthoscelides obtectus in cultivated common bean using a high-density genetic linkage map. BMC Plant Biol 22, 260.
DOI |
| [38] |
Macedo MLR, das Graças MachadoFreire M, da Silva MBR, Coelho LCB (2007). Insecticidal action of Bauhinia monandra leaf lectin (BmoLL) against Anagasta kuehniella (Lepidoptera: Pyralidae), Zabrotes subfasciatus and Callosobruchus maculatus (Coleoptera: Bruchidae). Comp Biochem Physiol A Mol Integr Physiol 146, 486-498.
DOI URL |
| [39] |
Mutungi C, Affognon HD, Njoroge AW, Manono J, Baributsa D, Murdock LL (2015). Triple-layer plastic bags protect dry common beans (Phaseolus vulgaris) against damage by Acanthoscelides obtectus (Coleoptera: Chrysomelidae) during storage. J Econ Entomol 108, 2479-2488.
DOI PMID |
| [40] |
Oliva MLV, Silva MCC, Sallai RC, Brito MV, Sampaio MU (2010). A novel subclassification for Kunitz proteinase in-hibitors from leguminous seeds. Biochimie 92, 1667-1673.
DOI URL |
| [41] |
Oliveira AS, Migliolo L, Aquino RO, Ribeiro JKC, Macedo LLP, Andrade LBS, Bemquerer MP, Santos EA, Kiyota S, de Sales MP (2007). Purification and characterization of a trypsin-papain inhibitor from Pithecelobium dumosum seeds and its in vitro effects towards digestive enzymes from insect pests. Plant Physiol Biochem 45, 858-865.
DOI URL |
| [42] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, MallerJ, Sklar P, Daly MJ, Sham PC (2007). PLINK: a tool set for whole-ge-nome association and population-based linkage analyses. Am J Hum Genet 81, 559-575.
DOI PMID |
| [43] |
Rakotoson T, Dusserre J, Letourmy P, Frouin J, Ratsimiala IR, Rakotoarisoa NV, Cao TV, Brocke KV, Ramanantsoanirina A, Ahmadi N, Raboin LM (2021). Genome-wide association study of nitrogen use efficiency and agronomic traits in upland rice. Rice Sci 28, 379-390.
DOI |
| [44] | Santino A, Valsasina B, Lioi L, Vitale A, Bollini R (1991). Bean (Phaseolus vulgaris L.) seed lectins: a novel elec-trophoretic variant of Arcelin. Plant Physiol 10, 7-11. |
| [45] |
Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu SQ, Song QJ, Chavarro C, Torres-Torres M, Geffroy V, Moghaddam SM, Gao DY, Abernathy B, Barry K, Blair M, Brick MA, Chovatia M, Gepts P, Goodstein DM, Gonzales M, Hellsten U, Hyten DL, Jia GF, Kelly JD, Kudrna D, Lee R, Richard MMS, Miklas PN, Osorno JM, Rodrigues J, Thareau V, Urrea CA, Wang M, Yu Y, Zhang M, Wing RA, Cregan PB, Rokhsar DS, Jackson SA (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46, 707-713.
DOI PMID |
| [46] |
Schroeder HE, Gollasch S, Moore A, Tabe LM, Craig S, Hardie DC, Chrispeels MJ, Spencer D, Higgins TJV (1995). Bean α-amylase inhibitor confers resistance to the pea weevil (Bruchus pisorum) in transgenic peas (Pisum sativum L.). Plant Physiol 107, 1233-1239.
PMID |
| [47] |
Shade RE, Pratt RC, Pomeroy MA (1987). Development and mortality of the bean weevil, Acanthoscelides obtec-tus (Coleoptera: Bruchidae), on mature seeds of tepary beans, Phaseolus acutifolius, and common beans, Phaseolus vulgaris. Environ Entomol 16, 1067-1070.
DOI URL |
| [48] | Smith MR, Rao IM (2021). Common bean. In: Sadras VO, Calderini DF, eds. Crop Physiology Case Histories for Major Crops. London: Academic Press. pp. 384-406. |
| [49] |
Velten G, Rott AS, Cardona C, Dorn S (2007). The inhibi-tory effect of the natural seed storage protein Arcelin on the development of Acanthoscelides obtectus. J Stored Prod Res 43, 550-557.
DOI URL |
| [50] |
Visscher PM, Brown MA, McCarthy MI, Yang J (2012). Five years of GWAS discovery. Am J Hum Genet 90, 7-24.
DOI PMID |
| [51] |
Wu J, Wang LF, Fu JJ, Chen JB, Wei SH, Zhang SL, Zhang J, Tang YS, Chen ML, Zhu JF, Lei L, Geng QH, Liu CL, Wu L, Li XM, Wang XL, Wang Q, Wang ZL, Xing SL, Zhang HK, Blair MW, Wang SM (2020). Resequencing of 683 common bean genotypes identifies yield component trait associations across a north-south cline. Nat Genet 52, 118-125.
DOI PMID |
| [52] | Yin LL, Zhang HH, Tang ZS, Xu JY, Yin D, Zhang ZW, Yuan XH, Zhu MJ, Zhao SH, Li XY, Liu XL (2021). rMVP: a memory-efficient, visualization-enhanced, and parallel- accelerated tool for genome-wide association study. Ge-nom Proteom Bioinf 19, 619-628. |
| [53] |
Yuste-Lisbona FJ, González AM, Capel C, García-Alcázar M, Capel J, De Ron AM, Lozano R, Santalla M (2014). Genetic analysis of single-locus and epistatic QTLs for seed traits in an adapted × nuña RIL population of common bean (Phaseolus vulgaris L.). Theor Appl Genet 127, 897-912.
DOI PMID |
| [54] |
Zaugg I, Magni C, Panzeri D, Daminati MG, Bollini R, Benrey B, Bacher S, Sparvoli F (2013). QUES, a new Phaseolus vulgaris genotype resistant to common bean weevils, contains the Arcelin-8 allele coding for new lectin-related variants. Theor Appl Genet 126, 647-661.
DOI URL |
| [1] | 崔娟, 于晓玉, 于跃娇, 梁铖玮, 孙健, 陈温福. 影响中国东北和日本粳稻食味品质差异的质构因素及其遗传基础解析[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 仝淼, 王欢, 张文双, 王超, 宋建潇. 重金属污染土壤中细菌抗生素抗性基因分布特征[J]. 生物多样性, 2025, 33(3): 24101-. |
| [3] | 武棒棒, 郝宇琼, 杨淑斌, 黄雨茜, 关攀锋, 郑兴卫, 赵佳佳, 乔玲, 李晓华, 刘维仲, 郑军. 山西小麦籽粒叶黄素含量变异及遗传特性分析[J]. 植物学报, 2023, 58(4): 535-547. |
| [4] | 吕善武, 张昌伟, 侯喜林, 邓书林. 白菜类作物抗TuMV研究进展[J]. 植物学报, 2023, 58(2): 335-346. |
| [5] | 李园, 常玉洁, 王兰芬, 王述民, 武晶. 普通菜豆镰孢菌枯萎病抗性种质资源筛选及全基因组关联分析[J]. 植物学报, 2023, 58(1): 51-61. |
| [6] | 张琦, 张文静, 袁宪凯, 李明, 赵强, 杜艳丽, 杜吉到. 褪黑素对盐胁迫下普通菜豆芽期核酸修复的调控机制[J]. 植物学报, 2023, 58(1): 108-121. |
| [7] | 谭文清, 陈军, 才宏伟. 黑麦草生物学研究进展[J]. 植物学报, 2022, 57(6): 802-813. |
| [8] | 金京波, 梁承志. 饲草基因组学研究进展[J]. 植物学报, 2022, 57(6): 732-741. |
| [9] | 陈孙禄, 詹成芳, 蒋红, 李琳涵, 张红生. 水稻籽粒灌浆速率的分子机制与遗传调控研究进展[J]. 植物学报, 2021, 56(1): 80-89. |
| [10] | 宣伟, 徐国华. 植物适应土壤氮素环境的基因选择: 以水稻为例[J]. 植物学报, 2021, 56(1): 1-5. |
| [11] | 赵宇慧, 李秀秀, 陈倬, 鲁宏伟, 刘羽诚, 张志方, 梁承志. 生物信息学分析方法I: 全基因组关联分析概述[J]. 植物学报, 2020, 55(6): 715-732. |
| [12] | 汪鸿儒, 储成才. 组学技术揭示水稻杂种优势遗传机制[J]. 植物学报, 2017, 52(1): 4-9. |
| [13] | 杨行海, 农保选, 夏秀忠, 张宗琼, 曾宇, 刘开强, 邓国富, 李丹婷. 水稻糯性相关基因的全基因组关联分析[J]. 植物学报, 2016, 51(6): 737-742. |
| [14] | 厉新民, 林鸿宣. 全基因组关联分析实现水稻粒型自然变异的分子解析[J]. 植物学报, 2016, 51(4): 411-415. |
| [15] | 周镕, 王波, 杨睿, 李书, 樊琳琳, 曾千春, 罗琼. 粳稻子预44中稻瘟病数量抗性位点分析[J]. 植物学报, 2015, 50(6): 691-698. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||