

植物学报 ›› 2023, Vol. 58 ›› Issue (2): 335-346.DOI: 10.11983/CBB22061 cstr: 32102.14.CBB22061
• 专题论坛 • 上一篇
收稿日期:2022-04-01
接受日期:2022-09-19
出版日期:2023-03-01
发布日期:2023-03-15
通讯作者:
*E-mail: 基金资助:
Shanwu Lü1, Changwei Zhang2, Xilin Hou2, Shulin Deng1,*( )
)
Received:2022-04-01
Accepted:2022-09-19
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: 摘要: 白菜类作物主要包括大白菜(Brassica rapa subsp. pekinensis)、不结球白菜(B. rapa subsp. chinensis)和芜菁(B. rapa subsp. rapa)等, 可用作蔬菜、油料及饲料。白菜类作物在生产过程中常受到病毒病的危害, 其中芜菁花叶病毒(TuMV)是主要病原物。近年来, 在TuMV抗性基因的遗传分析、定位克隆及分子机制方面取得了一定研究进展, 为培育优质抗病新品种奠定了基础。该文对国内外关于白菜类作物抗TuMV的遗传分析、抗性基因与分子标记、生物信息学与组学分析以及基因工程技术在抗TuMV研究中的应用进行综述, 以期为白菜类作物抗TuMV研究和技术利用提供科学依据。
吕善武, 张昌伟, 侯喜林, 邓书林. 白菜类作物抗TuMV研究进展. 植物学报, 2023, 58(2): 335-346.
Shanwu Lü, Changwei Zhang, Xilin Hou, Shulin Deng. Research Progress of TuMV Resistance for Brassica rapa. Chinese Bulletin of Botany, 2023, 58(2): 335-346.
| 抗性基因 | 植物材料 | 抗性品种 | 遗传/克隆情况 | TuMV株系 | 参考文献 |
|---|---|---|---|---|---|
| 显性基因/标记 | |||||
| ConTR01 | 大白菜 | RLR22 | 定位于R04, 候选基因可能是eIF(iso)4E | 1、3、4、7、 8、9和12 | Rusholme et al., |
| Tu1-Tu4 | 大白菜 | 91-112 | 4个QTLs, Tu1位于LG5连锁群, Tu2位于LG10连锁群, Tu3位于LG3连锁群, Tu4位于LG4连锁群 | C4 | Zhang et al., |
| TuR1-TuR4 | 大白菜 | A52-2 | TuR1和TuR2位于LG3连锁群, TuR3和TuR4位于LG7连锁群 | C3 | 张俊华等, |
| Tu1-Tu3 | 大白菜 | Y195-93 | Tu1位于R03连锁群, Tu2位于R04连锁群, Tu3位于R06连锁群 | C4 | 张晓伟等, |
| Rnt1-1 | 大白菜 | AS9 | 定位于R06连锁群, 与内源抗坏血酸和脱氢抗坏血酸合成相关 | UK1 | Fujiwara et al., |
| TuRBCH01 | 不结球白菜 | Q048 | 定位于R06连锁群 | C5 | Wang et al., |
| TuRB01b | 大白菜 | TD34-S1 | 定位于A06染色体, 与油菜TuRB01位于同一位点或互为等位基因 | UK1 | Lydiate et al., |
| TuRB07 | 大白菜 | VC1/VC40 | 定位于A06染色体, 候选基因是Bra018863, 属于CC-NBS-LRR基因 | C4 | Jin et al., |
| TuMV-R | 大白菜 | VC40 | 定位于A06染色体的0.34 Mb区域, 有6个候选基因, 4个CC-NBS-LRRs基因, 2个病程相关基因 | C4 | Chung et al., |
| TuRBCS01 | 大白菜 | 8407 | 定位于A04染色体的1.98 Mb区域 | C4 | Li et al., |
| 隐性基因/标记 | |||||
| retr01 | 大白菜 | RLR22 | 定位于R04连锁群, 候选基因可能是eIF(iso)4E | 1、3、4、7、 8、9和12 | Rusholme et al., |
| retr02 | 大白菜 | BP8407 | 定位于A04染色体, 候选基因是Bra035393, 编码eIF(iso)4E蛋白 | C4 | Qian et al., |
| trs | 大白菜 | SB18/SB22 | 定位于A04染色体, 可能与retr02紧密连锁或是其等位基因 | CHN2、CHN3、CHN4和CHN5 | Kim et al., |
| retrcs03 | 大白菜 | 73 | 定位于A04染色体, 候选基因是Bra032679, 编码γ-干扰素诱导蛋白 | C4 | 满卫萍, |
| 其它候选基因 | |||||
| BcTuR1 | 不结球白菜 | 短白梗 | 抗性候选基因, 受TuMV诱导高表达 | C4 | 马景蕃等, |
| BcTuR3 | 不结球白菜 | 短白梗 | 抗性候选基因, 受TuMV诱导高表达, 属于TIR- NB-LRR基因 | C4 | Ma et al., |
| BcLRK01 | 不结球白菜 | 泰三抗 | 抗性候选基因, 受TuMV诱导高表达, 属于LRR- RLKs基因 | C4 | 彭海涛等, |
| NhccGR1 | 不结球白菜 | 泰三抗 | 抗性候选基因, 受TuMV诱导高表达, 属于谷胱甘肽还原酶基因 | C4 | 李彦肖等, |
| BcTFIIIA | 不结球白菜 | NHCC001 | TuMV抗性基因, 编码锌指转录因子 | C4 | Zhang et al., |
表1 白菜抗TuMV基因及标记
Table 1 The TuMV resistance genes and makers in Brassica rapa
| 抗性基因 | 植物材料 | 抗性品种 | 遗传/克隆情况 | TuMV株系 | 参考文献 |
|---|---|---|---|---|---|
| 显性基因/标记 | |||||
| ConTR01 | 大白菜 | RLR22 | 定位于R04, 候选基因可能是eIF(iso)4E | 1、3、4、7、 8、9和12 | Rusholme et al., |
| Tu1-Tu4 | 大白菜 | 91-112 | 4个QTLs, Tu1位于LG5连锁群, Tu2位于LG10连锁群, Tu3位于LG3连锁群, Tu4位于LG4连锁群 | C4 | Zhang et al., |
| TuR1-TuR4 | 大白菜 | A52-2 | TuR1和TuR2位于LG3连锁群, TuR3和TuR4位于LG7连锁群 | C3 | 张俊华等, |
| Tu1-Tu3 | 大白菜 | Y195-93 | Tu1位于R03连锁群, Tu2位于R04连锁群, Tu3位于R06连锁群 | C4 | 张晓伟等, |
| Rnt1-1 | 大白菜 | AS9 | 定位于R06连锁群, 与内源抗坏血酸和脱氢抗坏血酸合成相关 | UK1 | Fujiwara et al., |
| TuRBCH01 | 不结球白菜 | Q048 | 定位于R06连锁群 | C5 | Wang et al., |
| TuRB01b | 大白菜 | TD34-S1 | 定位于A06染色体, 与油菜TuRB01位于同一位点或互为等位基因 | UK1 | Lydiate et al., |
| TuRB07 | 大白菜 | VC1/VC40 | 定位于A06染色体, 候选基因是Bra018863, 属于CC-NBS-LRR基因 | C4 | Jin et al., |
| TuMV-R | 大白菜 | VC40 | 定位于A06染色体的0.34 Mb区域, 有6个候选基因, 4个CC-NBS-LRRs基因, 2个病程相关基因 | C4 | Chung et al., |
| TuRBCS01 | 大白菜 | 8407 | 定位于A04染色体的1.98 Mb区域 | C4 | Li et al., |
| 隐性基因/标记 | |||||
| retr01 | 大白菜 | RLR22 | 定位于R04连锁群, 候选基因可能是eIF(iso)4E | 1、3、4、7、 8、9和12 | Rusholme et al., |
| retr02 | 大白菜 | BP8407 | 定位于A04染色体, 候选基因是Bra035393, 编码eIF(iso)4E蛋白 | C4 | Qian et al., |
| trs | 大白菜 | SB18/SB22 | 定位于A04染色体, 可能与retr02紧密连锁或是其等位基因 | CHN2、CHN3、CHN4和CHN5 | Kim et al., |
| retrcs03 | 大白菜 | 73 | 定位于A04染色体, 候选基因是Bra032679, 编码γ-干扰素诱导蛋白 | C4 | 满卫萍, |
| 其它候选基因 | |||||
| BcTuR1 | 不结球白菜 | 短白梗 | 抗性候选基因, 受TuMV诱导高表达 | C4 | 马景蕃等, |
| BcTuR3 | 不结球白菜 | 短白梗 | 抗性候选基因, 受TuMV诱导高表达, 属于TIR- NB-LRR基因 | C4 | Ma et al., |
| BcLRK01 | 不结球白菜 | 泰三抗 | 抗性候选基因, 受TuMV诱导高表达, 属于LRR- RLKs基因 | C4 | 彭海涛等, |
| NhccGR1 | 不结球白菜 | 泰三抗 | 抗性候选基因, 受TuMV诱导高表达, 属于谷胱甘肽还原酶基因 | C4 | 李彦肖等, |
| BcTFIIIA | 不结球白菜 | NHCC001 | TuMV抗性基因, 编码锌指转录因子 | C4 | Zhang et al., |
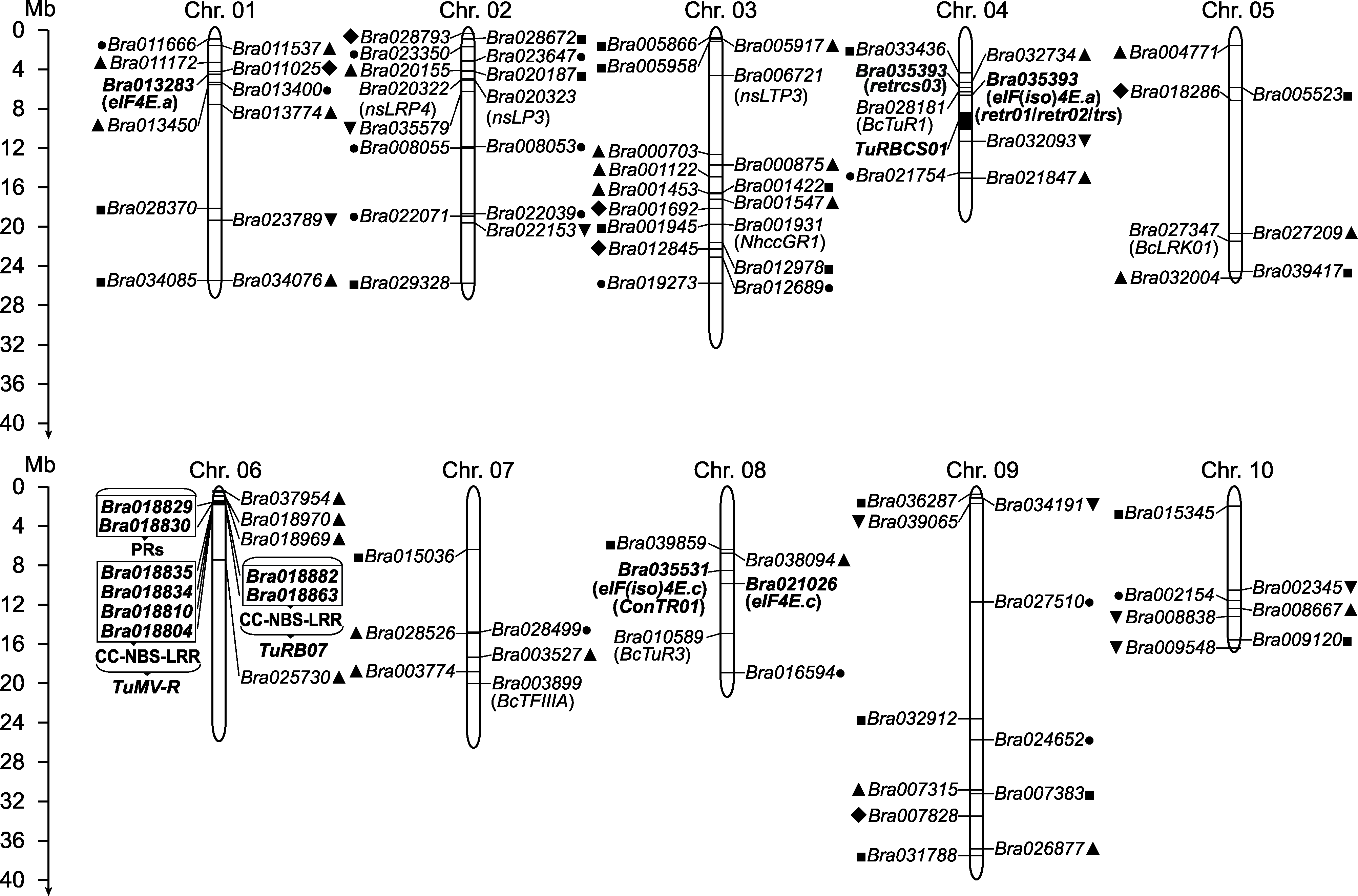
图1 TuMV抗性基因及位点在白菜染色体上的分布 黑色粗体表示通过分子标记筛选到的TuMV抗性相关基因或位点。黑色非粗体表示通过同源克隆、基因家族分析或组学分析筛选得到的候选基因。实心黑点和实心正方形分别表示通过对TIR-NBS-LRR家族和SNARE家族分析得到的候选基因; 实心正三角、实心倒三角形和实心菱形分别表示通过蛋白质组分析、全基因组关联分析和代谢组分析得到的候选基因。为便于分析, 根据最新注释将定位于Scaffold000104上的Bra035393定位到A04染色体的对应位置。本图基于Brara_Chiifu_V1.5基因组(Wang et al., 2011b)使用MG2C v2.1软件(Chao et al., 2021)绘制。
Figure 1 The distribution of TuMV resistance genes and loci on Chinese cabbage chromosomes The TuMV resistance genes and loci from the screening of molecular marks are highlighted in black bold font. The candidate genes from homology-based cloning, gene family analysis, and omics analysis are indicated in black non bold font. The candidate resistance genes from TIR-NBS-LRR and SNARE gene family analysis are marked separately by black dots and squares. The ones from proteome analysis, genome wide association study, and metabolome analysis are marked by black equilateral triangles, inverted triangles, and rhombus separately. Bra035393 from Scaffold000104 is updated to chromosome A04 according to the latest genome annotation. This distribution map is drawn based on Brara_Chiifu_V1.5 genome (Wang et al., 2011b) using MG2C v2.1 software (Chao et al., 2021).
| [1] | 曹光亮, 曹寿椿 (1995). 不结球白菜抗病育种研究.V. 抗TuMV遗传研究. 南京农业大学学报 18, 106-108. |
| [2] | 冯兰香, 徐玲, 刘佳, 钮心恪, 李秀生 (1988). 北京地区大白菜芜菁花叶病毒株系的鉴定. 中国蔬菜 (4), 11-13. |
| [3] | 高会超, 曾强, 张志刚, 赵智中, 裴玉贺, 刘栓桃, 李溢真, 刘贤娴, 宋希云, 李巧云 (2016). 与大白菜TuMV抗病基因TuRBCS01紧密连锁的分子标记开发. 农业生物技术学报 24, 196-205. |
| [4] | 国家蔬菜抗病育种课题TuMV株系研究协作组 (1989). 我国十省(市)十字花科蔬菜芜菁花叶病毒(TuMV)株系分化研究(II)——新鉴别寄主筛选及株系划分. 科学通报 (21), 1660-1664. |
| [5] | 韩和平 (2003). 大白菜抗芜菁花叶病毒病基因的AFLP分子标记的研究. 硕士论文. 北京: 中国农业科学院. pp. 3-60. |
| [6] |
李国亮, 钱伟, 张淑江, 李菲, 章时藩, 张慧, 谢露露, 武剑, 王晓武, 孙日飞 (2017). 白菜翻译起始因子eIF(iso)4E.a/c与芜菁花叶病毒TuMV-C4/UK1蛋白互作分析. 园艺学报 44, 1299-1308.
DOI |
| [7] |
李国亮, 张淑江, 钱伟, 李菲, 章时蕃, 张慧, 方智远, 孙日飞 (2019). 大白菜抗TuMV显性位点F2的QTL-seq分析. 园艺学报 46, 307-316.
DOI |
| [8] | 李树德 (1995). 中国主要蔬菜抗病育种进展. 北京: 科学出版社. pp. 33-34. |
| [9] | 李彦肖, 彭海涛, 管苇, 许小海, 李英, 侯喜林 (2012). 不结球白菜谷胱甘肽还原酶基因NhccGR1的克隆与表达分析. 南京农业大学学报 35(3), 7-12. |
| [10] | 刘琳 (2008). 不结球白菜芜菁花叶病毒病抗性机制及其遗传规律研究. 硕士论文. 南京: 南京农业大学. pp. 1-41. |
| [11] | 刘栓桃, 赵智中, 何启伟 (2004). 大白菜生物技术研究进展. 植物学通报 21, 709-718. |
| [12] | 刘栩平, 路文长, 刘元凯, 宋淑敏, 张成良, 王翠花 (1991). 芜青花叶病毒单克隆抗体杂交瘤细胞株的建立及对7个株系抗原决定簇的分析. 科学通报 (18), 1414-1417. |
| [13] |
刘勇, 李凡, 李月月, 张松柏, 高希武, 谢艳, 燕飞, 张安盛, 戴良英, 程兆榜, 丁铭, 牛颜冰, 王升吉, 车海彦, 江彤, 史晓斌, 何自福, 吴云锋, 张德咏, 青玲, 严婉荣, 杨学辉, 汤亚飞, 郑红英, 唐前君, 章松柏, 章东方, 蔡丽, 陶小荣 (2019). 侵染我国主要蔬菜作物的病毒种类、分布与发生趋势. 中国农业科学 52, 239-261.
DOI |
| [14] | 鹿英杰, 康永春, 李光池, 李东阁 (1988). 大白菜对TuMV抗性遗传规律的研究. 黑龙江农业科学 (6), 27-31. |
| [15] | 吕善武 (2019). TuMV侵染下的大白菜表达谱与蛋白组联合分析和TNL & SNARE基因分析验证. 博士论文. 南京: 南京农业大学. pp. 1-103. |
| [16] | 马景蕃, 史公军, 侯喜林, 孙菲菲, 冯岩, 吴海涛 (2009). 不结球白菜BcTuR1基因的克隆及表达. 西北植物学报 29, 2174-2180. |
| [17] | 满卫萍 (2020). 大白菜TuMV抗性基因retrcs03的定位. 硕士论文. 济南: 山东师范大学. pp. 1-42. |
| [18] | 满卫萍, 张志刚, 刘栓桃, 王荣花, 王立华, 许念芳, 徐文玲, 刘贤娴, 刘辰, 李巧云, 赵智中 (2019). 大白菜芜菁花叶病毒病抗性等位性研究. 农业生物技术学报 27, 2120-2129. |
| [19] | 钮心恪 (1984). 大白菜抗霜霉病, 病毒病原始材料的筛选及抗性遗传的研究. 中国蔬菜 (4), 28-32. |
| [20] | 彭海涛, 李彦肖, 李英, 侯喜林 (2012a). 不结球白菜BcLRK01基因对芜菁花叶病毒及胁迫诱导的响应. 西北植物学报 32, 670-675. |
| [21] | 彭海涛, 宋小明, 侯喜林, 李英 (2012b). 不结球白菜抗芜菁花叶病毒基因的遗传分析及SSR标记. 江苏农业学报 28, 396-401. |
| [22] | 钱伟, 张淑江, 章时蕃, 李菲, 张慧, 孙日飞 (2012). 大白菜TuMV抗性的主基因+多基因混合遗传分析. 中国蔬菜 (12), 16-21. |
| [23] | 屈淑平, 张彤, 张俊华, 崔崇士 (2009). 大白菜抗TuMV-C3的QTL分析. 东北农业大学学报 40(11), 33-37. |
| [24] | 佟容, 乔宏宇, 王智博, 吴春燕, 陈姗姗, 孙雨晴, 于占东 (2020a). 双价反义载体构建及对大白菜的遗传转化. 吉林农业大学学报 42, 533-537. |
| [25] | 佟容, 于占东, 吴春燕, 乔宏宇, 乔建磊, 王彧彧, 张新影 (2020b). 无选择标记转基因大白菜的检测. 吉林农业大学学报 42, 364-369. |
| [26] | 王新华 (2010). 不结球白菜抗芜菁花叶病毒基因分子标记与遗传定位. 博士论文 上海: 上海交通大学. pp. 1-58. |
| [27] | 汪影, 张昌伟, 吕善武, 侯喜林 (2018). 大白菜BrCNGC全基因组鉴定及其表达分析. 南京农业大学学报 41, 994-1002. |
| [28] | 吴斌, 张眉, 姜珊珊, 张安盛, 崔汉青, 辛志梅, 马立平, 王升吉 (2018). 大白菜、萝卜芜菁花叶病毒系统进化及CP序列分析. 山东农业科学 50(8), 100-105. |
| [29] | 阎瑾琦 (2000). 大白菜抗芜菁花叶病毒病基因的RAPD分子标记研究. 硕士论文. 北京: 中国农业科学院. pp. 1-48. |
| [30] | 杨学东, 戴薇, 张昌伟, 王金彦, 孔敏, 侯喜林 (2012). 白菜病毒诱导基因沉默技术体系的建立. 园艺学报 39, 2168-2174. |
| [31] | 于占东, 赵双宜, 何启伟, 王翠花, 牟晋华, 刘春香, 康静 (2006). TuMV-Nib反义基因对大白菜的遗传转化研究. 园艺学报 33, 1093-1095. |
| [32] | 张俊华, 屈淑平, 崔崇士 (2008). 大白菜抗芜菁花叶病毒的QTL分析. 植物病理学报 38, 178-184. |
| [33] | 张明, 潘春清, 张俊华, 屈淑萍, 崔崇士 (2011). 大白菜抗TuMV-C4的QTL分析. 中国瓜菜 24(2), 1-5. |
| [34] | 张晓伟, 原玉香, 王晓武, 孙日飞, 武剑, 谢从华, 蒋武生, 姚秋菊 (2009). 大白菜DH群体TuMV抗性的QTL定位与分析. 园艺学报 36, 731-736. |
| [35] | 朱常香, 宋云枝, 张松, 郭兴启, 温孚江 (2001). 抗芜菁花叶病毒转基因大白菜的培育. 植物病理学报 31, 257-264. |
| [36] |
Chao JT, Li ZY, Sun YH, Aluko OO, Wu XR, Wang Q, Liu GS (2021). MG2C: a user-friendly online tool for drawing genetic maps. Mol Horticulture 1, 16.
DOI |
| [37] | Chung H, Jeong YM, Mun JH, Lee SS, Chung WH, Yu HJ (2014). Construction of a genetic map based on high- throughput SNP genotyping and genetic mapping of a TuMV resistance locus in Brassica rapa. Mol Genet Genomics 289, 149-160. |
| [38] |
Domingo E, Holland JJ (1997). RNA virus mutations and fitness for survival. Annu Rev Microbiol 51, 151-178.
PMID |
| [39] |
Fujiwara A, Inukai T, Kim BM, Masuta C (2011). Combinations of a host resistance gene and the CI gene of turnip mosaic virus differentially regulate symptom expression in Brassica rapa cultivars. Arch Virol 156, 1575-1581.
DOI PMID |
| [40] |
Fujiwara A, Togawa S, Hikawa T, Matsuura H, Masuta C, Inukai T (2016). Ascorbic acid accumulates as a defense response to turnip mosaic virus in resistant Brassica rapa cultivars. J Exp Bot 67, 4391-4402.
DOI PMID |
| [41] |
Gardner MW, Kendrick JB (1921). Turnip mosaic. J Agric Res 22, 123-124.
DOI URL |
| [42] |
Green SK, Deng TC (1985). Turnip mosaic virus strains in cruciferous hosts in Taiwan. Plant Dis 69, 28-31.
DOI URL |
| [43] |
Hancock JM, Chaleeprom W, Chaleeprom W, Dale J, Gibbs A (1995). Replication slippage in the evolution of potyviruses. J Gen Virol 76, 3229-3232.
DOI URL |
| [44] |
Jenner CE, Keane GJ, Jones JE, Walsh JA (1999). Serotypic variation in turnip mosaic virus. Plant Pathol 48, 101-108.
DOI URL |
| [45] |
Jenner CE, Walsh JA (1996). Pathotypic variation in turnip mosaic virus with special reference to European isolates. Plant Pathol 45, 848-856.
DOI URL |
| [46] |
Jin MN, Lee SS, Ke L, Kim JS, Seo MS, Sohn SH, Park BS, Bonnema G (2014). Identification and mapping of a novel dominant resistance gene, TuRB07 to turnip mosaic virus in Brassica rapa. Theor Appl Genet 127, 509-519.
DOI URL |
| [47] | Kawakubo S, Gao FL, Li SF, Tan ZY, Huang YK, Adkar-Purushothama CR, Gurikar C, Maneechoat P, Chiemsombat P, Aye SS, Furuya N, Shevchenko O, Špak J, Škorić D, Ho SYW, Ohshima K (2021). Genomic analysis of the Brassica pathogen turnip mosaic potyvirus reveals its spread along the former trade routes of the Silk Road. Proc Natl Acad Sci USA 118, e2021221118. |
| [48] |
Kim J, Kang WH, Hwang J, Yang HB, Dosun K, Oh CS, Kang BC (2014). Transgenic Brassica rapa plants over- expressing eIF(iso)4E variants show broad-spectrum turnip mosaic virus (TuMV) resistance. Mol Plant Pathol 15, 615-626.
DOI URL |
| [49] |
Kim J, Kang WH, Yang HB, Park S, Jang CS, Yu HJ, Kang BC (2013). Identification of a broad-spectrum recessive gene in Brassica rapa and molecular analysis of the eIF4E gene family to develop molecular markers. Mol Breeding 32, 385-398.
DOI URL |
| [50] | Leung H, Williams PH (1983). Cytoplasmic male sterile Brassica campestris breeding lines with resistance to clubroot, turnip mosaic, and downy mildew. HortScience 8, 774-775. |
| [51] |
Li GL, Zhang SF, Li F, Zhang H, Zhang SJ, Zhao JJ, Sun RF (2021). Variability in the viral protein linked to the genome of turnip mosaic virus influences interactions with eIF(iso)4Es in Brassica rapa. Plant Pathol J 37, 47-56.
DOI URL |
| [52] |
Li QY, Tong HS, Zhang ZG, Zhao ZZ, Song XY (2011). Inheritance and development of EST-SSR marker associated with turnip mosaic virus resistance in Chinese cabbage. Can J Plant Sci 91, 707-715.
DOI URL |
| [53] |
Li QY, Zhang XL, Zeng Q, Zhang ZG, Liu ST, Pei YH, Wang SF, Liu XX, Xu WL, Fu WM, Zhao ZZ, Song XY (2015). Identification and mapping of a novel turnip mosaic virus resistance gene TuRBCS01 in Chinese cabbage (Brassica rapa L.). Plant Breed 134, 221-225.
DOI URL |
| [54] | Ling L, Yang JY (1940). A mosaic disease of rape and other cultivated crucifers in China. Phytopathology 30, 338-342. |
| [55] |
Liu C, Sun GS, Zhang RJ, Lv SW, Gao L, Gao LW, Liu TK, Xiao D, Hou XL, Zhang CW (2019). Proteomic analysis provides integrated insight into mechanisms of turnip mosaic virus long distance movement in Brassica rapa. Biol Plant 63, 164-173.
DOI URL |
| [56] | Liu ST, Li QY, Zhang ZG, Wang LH, Wang X, Zhao ZZ, Wang SF, Xu WL, Liu XX, Liu C (2017). Characterization of a double mutation of eIF(iso)4E.a and eIF(iso)4E.c with a diverse genetic background and TuMV resistance in Chinese cabbage (Brassica rapa ssp. pekinensis) J Agric Biotech 25, 1575-1587. |
| [57] |
Liu XP, Lu CW, Liu YK, Li JL (1990). A study on TuMV strain differentiation of cruciferous vegetables from ten provinces in China——new host differentiator screening and strain classification. Chin Sci Bull 35, 1734-1739.
DOI URL |
| [58] |
Lu XX, Zhang L, Huang WY, Zhang SJ, Zhang SF, Li F, Zhang H, Sun RF, Zhao JJ, Li GL (2022). Integrated volatile metabolomics and transcriptomics analyses reveal the influence of infection TuMV to volatile organic compounds in Brassica rapa. Horticulturae 8, 57.
DOI URL |
| [59] |
Lu Y, Dai SY, Gu A, Liu MY, Wang YH, Luo SX, Zhao YJ, Wang S, Xuan SX, Chen XP, Li XF, Bonnema G, Zhao JJ, Shen SX (2016). Microspore induced doubled haploids production from ethyl methanesulfonate (EMS) soaked flower buds is an efficient strategy for mutagenesis in Chinese cabbage. Front Plant Sci 7, 1780.
DOI PMID |
| [60] |
Lv SW, Zhang CW, Tang J, Li YX, Wang Z, Jiang DH, Hou XL (2015). Genome-wide analysis and identification of TIR-NBS-LRR genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveal expression patterns to TuMV infection. Physiol Mol Plant Pathol 90, 89-97.
DOI URL |
| [61] |
Lydiate DJ, Pilcher RLR, Higgins EE, Walsh JA (2014). Genetic control of immunity to turnip mosaic virus (TuMV) pathotype 1 in Brassica rapa (Chinese cabbage). Genome 57, 419-425.
DOI PMID |
| [62] | Lyu S, Gao LW, Zhang RJ, Zhang CW, Hou XL (2020). Correlation analysis of expression profile and quantitative iTRAQ-LC-MS/MS proteomics reveals resistance mechanism against TuMV in Chinese cabbage (Brassica rapa ssp. pekinensis). Front Genet 11, 963. |
| [63] |
Ma JF, Hou XL, Xiao D, Qi L, Wang F, Sun FF, Wang Q (2010). Cloning and characterization of the BcTuR3 gene related to resistance to turnip mosaic virus (TuMV) from non-heading Chinese cabbage. Plant Mol Biol Rep 28, 588-596.
DOI URL |
| [64] |
McDonald JG, Hiebert E (1974). Ultrastructure of cylindrical inclusions induced by viruses of the potato Y group as visualized by freeze-etching. Virology 58, 200-208.
PMID |
| [65] | Nagaharu U (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7, 389-452. |
| [66] |
Nellist CF, Ohshima K, Ponz F, Walsh JA (2022). Turnip mosaic virus, a virus for all seasons. Ann Appl Biol 180, 312-327.
DOI URL |
| [67] |
Nellist CF, Qian W, Jenner CE, Moore JD, Zhang SJ, Wang XW, Briggs WH, Barker GC, Sun RF, Walsh JA (2014). Multiple copies of eukaryotic translation initiation factors in Brassica rapa facilitate redundancy, enabling diversification through variation in splicing and broad-spect- rum virus resistance. Plant J 77, 261-268.
DOI URL |
| [68] |
Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, Tan ZY, Sano T, Azuhata F, Walsh JA, Fletcher J, Chen J, Gera A, Gibbs A (2002). Molecular evolution of turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol 83, 1511-1521.
DOI PMID |
| [69] |
Park CJ, Shin R, Park JM, Lee GJ, You JS, Paek KH (2002). Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol Biol 48, 243-254.
DOI URL |
| [70] | Provvidenti R (1980). Evaluation of Chinese cabbage cultivars from Japan and the People’s Republic of China for resistance to turnip mosaic virus and cauliflower mosaic virus. J Amer Soc Hortic Sci 105, 571-573. |
| [71] |
Pyott DE, Sheehan E, Molnar A (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol 17, 1276-1288.
DOI URL |
| [72] |
Qian W, Zhang SJ, Zhang SF, Li F, Zhang H, Wu J, Wang XW, Walsh JA, Sun RF (2013). Mapping and candidate-gene screening of the novel turnip mosaic virus resistance gene retr02 in Chinese cabbage (Brassica rapa L.). Theor Appl Genet 126, 179-188.
DOI PMID |
| [73] |
Robaglia C, Caranta C (2006). Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11, 40-45.
DOI PMID |
| [74] |
Rusholme RL, Higgins EE, Walsh JA, Lydiate DJ (2007). Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J Gen Virol 88, 3177-3186.
DOI PMID |
| [75] |
Sánchez F, Wang X, Jenner CE, Walsh JA, Ponz F (2003). Strains of turnip mosaic potyvirus as defined by the molecular analysis of the coat protein gene of the virus. Virus Res 94, 33-43.
PMID |
| [76] |
Sarowar S, Kim YJ, Kim KD, Hwang BK, Ok SH, Shin JS (2009). Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep 28, 419-427.
DOI PMID |
| [77] |
Smith KM (1935). A virus disease of cultivated crucifers. Ann Appl Biol 22, 239-242.
DOI URL |
| [78] |
Suh SK, Green SK, Park HG (1995). Genetics of resistance to five strains of turnip mosaic virus in Chinese cabbage. Euphytica 81, 71-77.
DOI URL |
| [79] |
Sun XX, Li X, Lu Y, Wang S, Zhang XM, Zhang K, Su XJ, Liu MY, Feng DL, Luo SX, Gu AX, Fu Y, Chen XP, Xuan SX, Wang YH, Xu DH, Chen SM, Ma W, Shen SX, Cheng F, Zhao JJ (2022). Construction of a high-density mutant population of Chinese cabbage facilitates the genetic dissection of agronomic traits. Mol Plant 15, 913-924.
DOI URL |
| [80] |
The Brassica Rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43, 1035-1039.
DOI |
| [81] |
Tomimura K, Gibbs AJ, Jenner CE, Walsh JA, Ohshima K (2003). The phylogeny of turnip mosaic virus; compa- risons of 38 genomic sequences reveal a Eurasian origin and a recent ‘emergence’ in east Asia. Mol Ecol 12, 2099-2111.
PMID |
| [82] |
Walsh JA, Jenner CE (2002). Turnip mosaic virus and the quest for durable resistance. Mol Plant Pathol 3, 289-300.
DOI PMID |
| [83] | Wang N, Zhang Y, Huang SN, Liu ZY, Li CY, Feng H (2020). Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hort Res 7, 8. |
| [84] |
Wang XH, Yang L, Chen HY (2011). A linkage map of Pak-choi (Brassica rapa ssp. chinensis) based on AFLP and SSR markers and identification of AFLP markers for resistance to TuMV. Plant Breed 130, 275-277.
DOI URL |
| [85] |
Wang Z, Jiang DH, Zhang CW, Tan HW, Li YX, Lv SW, Hou XL, Cui XY (2015). Genome-wide identification of turnip mosaic virus-responsive microRNAs in non-heading Chinese cabbage by high-throughput sequencing. Gene 571, 178-187.
DOI PMID |
| [86] | Wei TY, Zhang CW, Hou XL, Sanfaçon H, Wang AM (2013). The SNARE protein Syp71 is essential for turnip mo- saic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog 9, e1003378. |
| [87] |
Yoon JY, Green SK, Opena RT (1993). Inheritance of resistance to turnip mosaic virus in Chinese cabbage. Euphytica 69, 103-108.
DOI URL |
| [88] |
Yoshii H (1963). On the strain distribution of turnip mosaic virus. Jpn J Phytopathol 28, 221-227.
DOI URL |
| [89] |
Yu J, Gao LW, Yang Y, Liu C, Zhang RJ, Sun FF, Song LX, Xiao D, Liu TK, Hou XL, Zhang CW (2019). The methylation pattern of DNA and complex correlations with gene expressions during TuMV infection in Chinese cabbage. Biol Plant 63, 671-680.
DOI URL |
| [90] |
Yu J, Yang XD, Wang Q, Gao LW, Yang Y, Xiao D, Liu TK, Li Y, Hou XL, Zhang CW (2018). Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol Plant 62, 826-834.
DOI |
| [91] |
Zhang CW, Lyu S, Gao LW, Song XM, Li YX, Hou XL (2018). Genome-wide identification, classification, and expression analysis of SNARE genes in Chinese cabbage (Brassica rapa ssp. pekinensis) infected by turnip mosaic virus. Plant Mol Biol Rep 36, 210-224.
DOI |
| [92] | Zhang FL, Wang M, Liu XC, Zhao XY, Yang JP (2008). Quantitative trait loci analysis for resistance against turnip mosaic virus based on a doubled-haploid population in Chinese cabbage. Plant Breed 127, 82-86. |
| [93] | Zhang RJ, Liu C, Song XM, Sun FF, Xiao D, Wei YP, Hou XL, Zhang CW (2020). Genome-wide association study of turnip mosaic virus resistance in non-heading Chinese cabbage. 3 Biotech 10, 363. |
| [94] | Zhang RJ, Zhang CW, Lyu S, Fang ZY, Zhu HF, Hou XL (2022a). Functional analysis of BcSNX3 in regulating resistance to turnip mosaic virus (TuMV) by autophagy in Pak-choi (Brassica campestris ssp. chinensis). Agronomy 12, 1757. |
| [95] | Zhang RJ, Zhang CW, Lyu S, Wu HY, Yuan MG, Fang ZY, Li FF, Hou XL (2022b). BcTFIIIA negatively regulates turnip mosaic virus infection through interaction with viral CP and VPg proteins in Pak Choi (Brassica campestris ssp. chinensis). Genes 13, 1209. |
| [1] | 仝淼, 王欢, 张文双, 王超, 宋建潇. 重金属污染土壤中细菌抗生素抗性基因分布特征[J]. 生物多样性, 2025, 33(3): 24101-. |
| [2] | 顾红雅, 陈凡, 林荣呈, 漆小泉, 杨淑华, 陈之端, 陈学伟, 丁兆军, 萧浪涛, 左建儒, 姜里文, 白永飞, 种康, 王雷. 2024年中国植物科学重要研究进展[J]. 植物学报, 2025, 60(2): 151-171. |
| [3] | 耿江天, 王菲, 赵华斌. 城市化对中国蝙蝠影响的研究进展[J]. 生物多样性, 2024, 32(8): 24109-. |
| [4] | 刘荆州, 钱易鑫, 张燕雪丹, 崔凤. 基于潜在迪利克雷分布(LDA)模型的旗舰物种范式研究进展与启示[J]. 生物多样性, 2024, 32(4): 23439-. |
| [5] | 陈凡, 顾红雅, 漆小泉, 林荣呈, 钱前, 萧浪涛, 杨淑华, 左建儒, 白永飞, 陈之端, 丁兆军, 王小菁, 姜里文, 种康, 王雷. 2023年中国植物科学重要研究进展[J]. 植物学报, 2024, 59(2): 171-187. |
| [6] | 杨淑华, 钱前, 左建儒, 顾红雅, 漆小泉, 林荣呈, 陈凡, 王小菁, 萧浪涛, 白永飞, 姜里文, 王雷, 陈之端, 种康, 王台. 2022年中国植物科学重要研究进展[J]. 植物学报, 2023, 58(2): 175-188. |
| [7] | 罗瑞, 陈娅, 张汉马. 芸薹属植物全基因组重测序研究进展[J]. 生物多样性, 2023, 31(10): 23237-. |
| [8] | 李晓明, 王兰芬, 唐永生, 常玉洁, 张菊香, 王述民, 武晶. 普通菜豆抗菜豆象性状的全基因组关联分析[J]. 植物学报, 2023, 58(1): 77-89. |
| [9] | 李园, 常玉洁, 王兰芬, 王述民, 武晶. 普通菜豆镰孢菌枯萎病抗性种质资源筛选及全基因组关联分析[J]. 植物学报, 2023, 58(1): 51-61. |
| [10] | 葛颂. 中国植物系统和进化生物学研究进展[J]. 生物多样性, 2022, 30(7): 22385-. |
| [11] | 杨凯如, 贾绮玮, 金佳怡, 叶涵斐, 王盛, 陈芊羽, 管易安, 潘晨阳, 辛德东, 方媛, 王跃星, 饶玉春. 水稻黄绿叶调控基因YGL18的克隆与功能解析[J]. 植物学报, 2022, 57(3): 276-287. |
| [12] | 陈凡, 顾红雅, 漆小泉, 钱前, 左建儒, 杨淑华, 陈之端, 王雷, 林荣呈, 姜里文, 王小菁, 萧浪涛, 白永飞, 种康, 王台. 2021年中国植物科学重要研究进展[J]. 植物学报, 2022, 57(2): 139-152. |
| [13] | 江建华, 党小景, 姚文豪, 胡梦竹, 王雨停, 胡长敏, 张瑛, 王德正. 水稻核不育系4个柱头性状的遗传分析[J]. 植物学报, 2021, 56(3): 284-295. |
| [14] | 顾红雅, 左建儒, 漆小泉, 杨淑华, 陈之端, 钱前, 林荣呈, 王雷, 萧浪涛, 王小菁, 陈凡, 姜里文, 白永飞, 种康, 王台. 2020年中国植物科学若干领域重要研究进展[J]. 植物学报, 2021, 56(2): 119-133. |
| [15] | 井新, 贺金生. 生物多样性与生态系统多功能性和多服务性的关系: 回顾与展望[J]. 植物生态学报, 2021, 45(10): 1094-1111. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||