

植物学报 ›› 2023, Vol. 58 ›› Issue (2): 175-188.DOI: 10.11983/CBB23061 cstr: 32102.14.CBB23061
• 主编评述 • 下一篇
杨淑华1, 钱前2, 左建儒3, 顾红雅4, 漆小泉5, 林荣呈5, 陈凡3, 王小菁6, 萧浪涛7, 白永飞5, 姜里文8, 王雷5, 陈之端5, 种康5,*( ), 王台5,*(
), 王台5,*( )
)
出版日期:2023-03-01
发布日期:2023-05-12
Shuhua Yang1, Qian Qian2, Jianru Zuo3, Hongya Gu4, Xiaoquan Qi5, Rongcheng Lin5, Fan Chen3, Xiaojing Wang6, Xiao Langtao7, Yongfei Bai5, Liwen Jiang8, Lei Wang5, Zhiduan Chen5, Kang Chong5,*( ), Tai Wang5,*(
), Tai Wang5,*( )
)
Online:2023-03-01
Published:2023-05-12
About author:E-mail: 摘要: 2022年中国科学家在植物科学主流期刊发表的论文数量相比2021年显著增加, 在防止“多精受精”分子机制, 胞外pH感受器, 叶绿体蛋白运输通道结构, 作物高产、优质、耐逆、抗病及共生固氮机制, 被子植物自交不亲和性的起源和演化机制, 甘蔗和玉蜀黍种质资源演化等方面取得了重要研究进展。其中, “水稻抗高温基因挖掘及调控新机制”入选2022年度中国生命科学十大进展。该文总结了2022年度我国植物科学研究取得的成果, 简要介绍了30项有代表性的重要进展, 梳理了植物科学研究中所使用的实验材料, 以帮助读者了解我国植物科学的发展态势, 进而思考如何更好地开展下阶段研究和服务国家需求。
杨淑华, 钱前, 左建儒, 顾红雅, 漆小泉, 林荣呈, 陈凡, 王小菁, 萧浪涛, 白永飞, 姜里文, 王雷, 陈之端, 种康, 王台. 2022年中国植物科学重要研究进展. 植物学报, 2023, 58(2): 175-188.
Shuhua Yang, Qian Qian, Jianru Zuo, Hongya Gu, Xiaoquan Qi, Rongcheng Lin, Fan Chen, Xiaojing Wang, Xiao Langtao, Yongfei Bai, Liwen Jiang, Lei Wang, Zhiduan Chen, Kang Chong, Tai Wang. Achievements and Advances of Plant Sciences Research in China in 2022. Chinese Bulletin of Botany, 2023, 58(2): 175-188.
| 2020年 | 2021年 | 2022年 | ||||
|---|---|---|---|---|---|---|
| 文章 数量 | 占比 (%) | 文章 数量 | 占比 (%) | 文章 数量 | 占比 (%) | |
| 中国 | 589 | 43.6 | 640 | 44.1 | 742 | 50.3 |
| 美国 | 450 | 33.3 | 453 | 31.2 | 434 | 29.4 |
| 德国 | 229 | 17.0 | 239 | 16.5 | 205 | 13.9 |
| 英国 | 130 | 9.6 | 138 | 9.5 | 136 | 9.2 |
| 法国 | 115 | 8.5 | 114 | 7.9 | 104 | 7.0 |
表1 2020-2022年中国与4个欧美国家的科学家在5种植物科学主流期刊(MP、NP、PC、PP和PJ)的发文量比较(数据来源: Web of Science核心合集)
Table 1 The numbers of papers published by Scientists from China, America, Germany, UK and France in the five major journals of plant sciences (MP, NP, PC, PP and PJ) from 2020 to 2022 (data sources: Web of Science)
| 2020年 | 2021年 | 2022年 | ||||
|---|---|---|---|---|---|---|
| 文章 数量 | 占比 (%) | 文章 数量 | 占比 (%) | 文章 数量 | 占比 (%) | |
| 中国 | 589 | 43.6 | 640 | 44.1 | 742 | 50.3 |
| 美国 | 450 | 33.3 | 453 | 31.2 | 434 | 29.4 |
| 德国 | 229 | 17.0 | 239 | 16.5 | 205 | 13.9 |
| 英国 | 130 | 9.6 | 138 | 9.5 | 136 | 9.2 |
| 法国 | 115 | 8.5 | 114 | 7.9 | 104 | 7.0 |
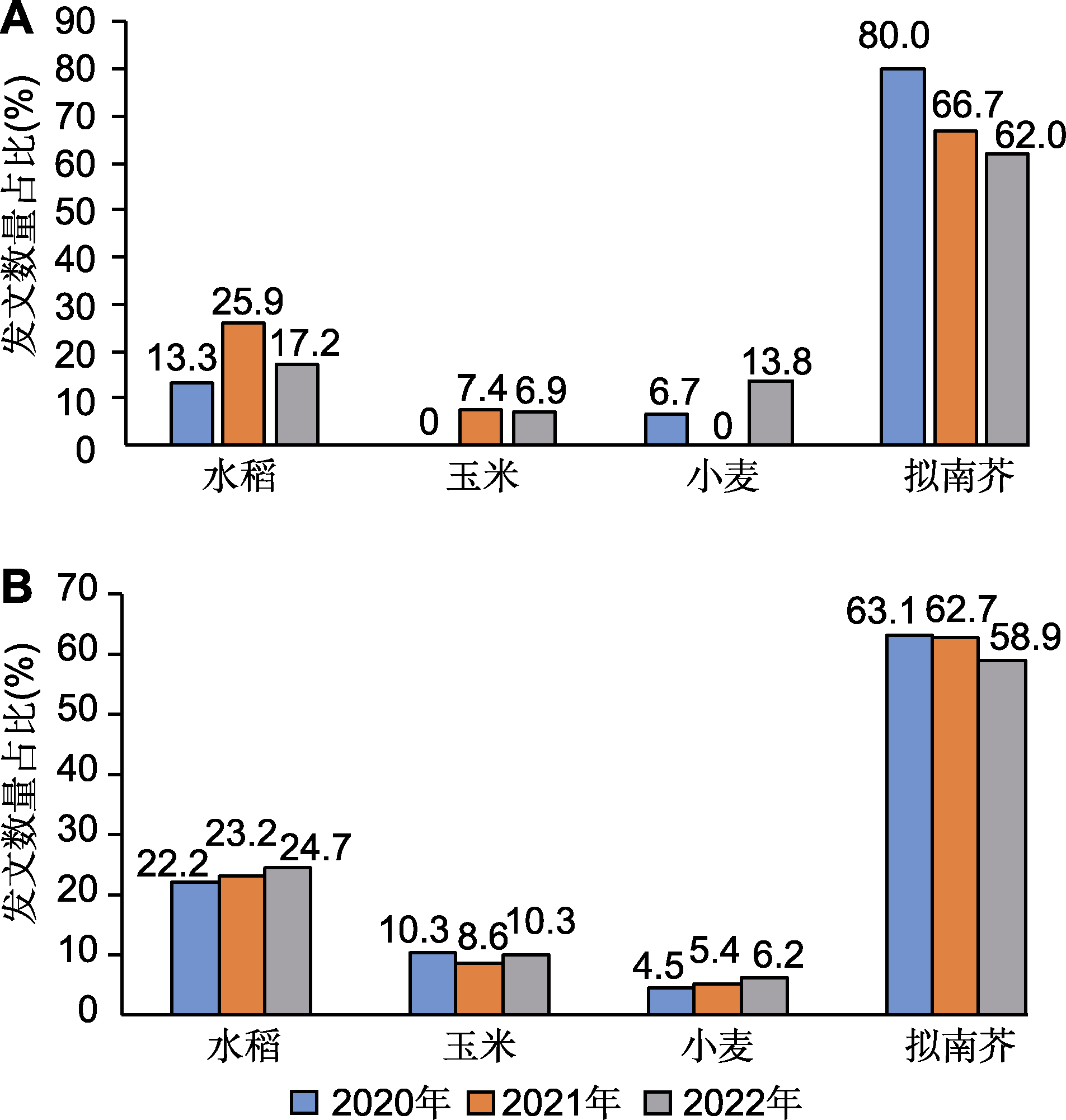
图1 2020-2022年中国植物科学家在国际综合性学术期刊(Science、Nature和Cell) (A)和植物科学主流期刊(MP、NP、PC、PP和PJ) (B)上发表以水稻、玉米、小麦和拟南芥为研究材料的论文数量占比(数据来源: Web of Science核心合集) 注: 以2个或2个以上物种为材料的文章被重复计数。3种国际综合性学术期刊(Science、Nature和Cell)刊登的以水稻、玉米、小麦和拟南芥为实验材料的文章数量总和分别为15 (2020年)、27 (2021年)和29 (2022年)篇; 5种(MP、NP、PC、PP和PJ)植物科学主流期刊刊登的以水稻、玉米、小麦和拟南芥为实验材料的文章数量总和分别为555 (2020年)、569 (2021年)和681 (2022年)。MP、NP、PC、PP和PJ同表1。
Figure 1 The proportion of papers published by scientists from China in international multidisciplinary journals (Science, Nature and Cell) (A) and mainstream plant science journals (MP, NP, PC, PP and PJ) (B) from 2020 to 2022 using rice, maize, wheat and Arabidopsis as research materials (data sources: Web of Science) Note: When two or more species are used as research materials in a paper, this paper is counted more than once accordingly. The total number of papers published in the three international multidisciplinary journals (Science, Nature and Cell) using rice, maize, wheat and Arabidopsis as research materials was 15 in 2020, 27 in 2021, and 29 in 2022, respectively. The total number of papers published in the five mainstream plant science journals (MP, NP, PC, PP and PJ) using rice, maize, wheat and Arabidopsis as research materials was 555 in 2020, 569 in 2021, and 681 in 2022, respectively. MP, NP, PC, PP and PJ are the same as shown in Table 1.
| [1] |
孔令让 (2022). 另辟蹊径破解小麦条锈病的基因密码. 植物学报 57, 405-408.
DOI |
| [2] |
杨永青, 郭岩 (2022). 植物细胞质外体pH感受机制的解析. 植物学报 57, 409-411.
DOI |
| [3] |
余泓, 李家洋 (2022). 是金子无论在何处都发光: 玉米和水稻驯化中的趋同选择. 植物学报 57, 153-156.
DOI |
| [4] | Abas L, Kolb M, Stadlmann J, Janacek DP, Lukic K, Schwechheimer C, Sazanov LA, Mach L, Friml J, Hammes UZ (2021). Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc Natl Acad Sci USA 118, e2020857118. |
| [5] |
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695-705.
DOI PMID |
| [6] |
Chen L, Luo JY, Jin ML, Yang N, Liu XG, Peng Y, Li WQ, Phillips A, Cameron B, Bernal JS, Rellán-Álvarez R, Sawers RJH, Liu Q, Yin YJ, Ye XN, Yan JL, Zhang QH, Zhang XT, Wu SS, Gui ST, Wei WJ, Wang YB, Luo Y, Jiang CL, Deng M, Jin M, Jian LM, Yu YH, Zhang ML, Yang XH, Hufford MB, Fernie AR, Warburton ML, Ross-Ibarra J, Yan JB (2022a). Genome sequencing reveals evidence of adaptive variation in the genus Zea. Nat Genet 54, 1736-1745.
DOI |
| [7] |
Chen PF, Liu X, Gu CH, Zhong PY, Song N, Li MB, Dai ZQ, Fang XQ, Liu ZM, Zhang JF, Tang RK, Fan SW, Lin XF (2022b). A plant-derived natural photosynthetic system for improving cell anabolism. Nature 612, 546-554.
DOI |
| [8] | Chen WK, Chen L, Zhang X, Yang N, Guo JH, Wang M, Ji SH, Zhao XY, Yin PF, Cai LC, Xu J, Zhang LL, Han YJ, Xiao YN, Xu G, Wang YB, Wang SH, Wu S, Yang F, Jackson D, Cheng JK, Chen SH, Sun CQ, Qin F, Tian F, Fernie AR, Li JS, Yan JB, Yang XH (2022c). Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375, eabg7985. |
| [9] |
Chen WQ, Wellings C, Chen XM, Kang ZS, Liu TG (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici Mol Plant Pathol 15, 433-446.
DOI URL |
| [10] |
Doebley JF, Gaut BS, Smith BD (2006). The molecular genetics of crop domestication. Cell 127, 1309-1321.
DOI PMID |
| [11] |
Felix G, Duran JD, Volko S, Boller T (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18, 265-276.
DOI PMID |
| [12] |
Feng XQ, Zilberman D, Dickinson H (2013). A conversation across generations: soma-germ cell crosstalk in plants. Dev Cell 24, 215-225.
DOI PMID |
| [13] |
Fujii S, Kubo KI, Takayama S (2016). Non-self- and self-recognition models in plant self-incompatibility. Nat Plants 2, 16130.
DOI PMID |
| [14] |
Gao J, Zhang K, Cheng YJ, Yu S, Shang GD, Wang FX, Wu LY, Xu ZG, Mai YX, Zhao XY, Zhai D, Lian H, Wang JW (2022). A robust mechanism for resetting juvenility during each generation in Arabidopsis. Nat Plants 8, 257-268.
DOI |
| [15] |
Gao ZY, Wang YF, Chen G, Zhang AP, Yang SL, Shang LG, Wang DY, Ruan BP, Liu CL, Jiang HZ, Dong GJ, Zhu L, Hu J, Zhang GH, Zeng DL, Guo LB, Xu GH, Teng S, Harberd NP, Qian Q (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat Commun 10, 5207.
DOI |
| [16] |
Huang C, Chen Z, Liang CZ (2021). Oryza pan-genomics: a new foundation for future rice research and improvement. Crop J 9, 622-632.
DOI |
| [17] | Huang GQ, Kilic A, Karady M, Zhang J, Mehra P, Song XY, Sturrock CJ, Zhu WW, Qin H, Hartman S, Schneider HM, Bhosale R, Dodd IC, Sharp RE, Huang RF, Mooney SJ, Liang WQ, Bennett MJ, Zhang DB, Pandey BK (2022a). Ethylene inhibits rice root elongation in compacted soil via ABA- and auxin-mediated mechanisms. Proc Natl Acad Sci USA 119, e2201072119. |
| [18] | Huang SJ, Jia AL, Song W, Hessler G, Meng YG, Sun Y, Xu LN, Laessle H, Jirschitzka J, Ma SC, Xiao Y, Yu DL, Hou J, Liu RQ, Sun HH, Liu XH, Han ZF, Chang JB, Parker JE, Chai JJ (2022b). Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science 377, eabq3297. |
| [19] |
Huang YC, Wang HH, Zhu YD, Huang X, Li S, Wu XG, Zhao Y, Bao ZG, Qin L, Jin YB, Cui YH, Ma GJ, Xiao Q, Wang Q, Wang JC, Yang XR, Liu HJ, Lu XD, Larkins BA, Wang WQ, Wu YR (2022c). THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 612, 292-300.
DOI |
| [20] |
Huffaker A, Pearce G, Ryan CA (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103, 10098-10103.
DOI PMID |
| [21] |
Hufford MB, Bilinski P, Pyhäjärvi T, Ross-Ibarra J (2012a). Teosinte as a model system for population and ecological genomics. Trends Genet 28, 606-615.
DOI URL |
| [22] | Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia JM, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, Lai JS, Morrell PL, Shannon LM, Song C, Springer NM, Swanson-Wagner RA, Tiffin P, Wang J, Zhang GY, Doebley J, McMullen MD, Ware D, Buckler ES, Yang S, Ross-Ibarra J (2012b). Comparative population genomics of maize domestication and improvement. Nat Genet 44, 808-811. |
| [23] | Ishida Y, Tsunashima M, Hiei Y, Komari T (2015). Wheat (Triticum aestivum L.) transformation using immature embryos. In: Wang K, ed. Agrobacterium Protocols. New York: Springer. pp. 189-198. |
| [24] |
Jamieson PA, Shan LB, He P (2018). Plant cell surface molecular cypher: receptor-like proteins and their roles in immunity and development. Plant Sci 274, 242-251.
DOI PMID |
| [25] | Jia AL, Huang SJ, Song W, Wang JL, Meng YG, Sun Y, Xu LN, Laessle H, Jirschitzka J, Hou J, Zhang TT, Yu WQ, Hessler G, Li ET, Ma SC, Yu DL, Gebauer J, Baumann U, Liu XH, Han ZF, Chang JB, Parker JE, Chai JJ (2022). TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science 377, eabq8180. |
| [26] |
Jiang HF, Shi YT, Liu JY, Li Z, Fu DY, Wu SF, Li MZ, Yang ZJ, Shi YL, Lai JS, Yang XH, Gong ZZ, Hua J, Yang SH (2022). Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat Plants 8, 1176-1190.
DOI PMID |
| [27] |
Jin ZY, Wan L, Zhang YQ, Li XC, Cao Y, Liu HB, Fan SY, Cao D, Wang ZM, Li XB, Pan JM, Dong MQ, Wu JP, Yan Z (2022). Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes. Cell 185, 4788-4800.
DOI URL |
| [28] |
Jones JDG, Dangl JL (2006). The plant immune system. Nature 444, 323-329.
DOI |
| [29] |
Kan Y, Mu XR, Zhang H, Gao J, Shan JX, Ye WW, Lin HX (2022). TT2 controls rice thermotolerance through SCT1- dependent alteration of wax biosynthesis. Nat Plants 8, 53-67.
DOI PMID |
| [30] |
Ke XL, Xiao H, Peng YQ, Wang J, Lv Q, Wang XL (2022). Phosphoenolpyruvate reallocation links nitrogen fixation rates to root nodule energy state. Science 378, 971-977.
DOI PMID |
| [31] |
Langner T, Kamoun S, Belhaj K (2018). CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol 56, 479-512.
DOI PMID |
| [32] |
Li SN, Lin DX, Zhang YW, Deng M, Chen YX, Lv B, Li BS, Lei Y, Wang YP, Zhao L, Liang YT, Liu JX, Chen KL, Liu ZY, Xiao J, Qiu JL, Gao CX (2022a). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455-460.
DOI |
| [33] |
Li XM, Chao DY, Wu Y, Huang XH, Chen K, Cui LG, Su L, Ye WW, Chen H, Chen HC, Dong NQ, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan JX, Gao JP, Lin HX (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat Genet 47, 827-833.
DOI |
| [34] | Li Y, Du YX, Huai JL, Jing YJ, Lin RC (2022b). The RNA helicase UAP56 and the E3 ubiquitin ligase COP1 coordinately regulate alternative splicing to repress photomorphogenesis in Arabidopsis. Plant Cell 34, 4191-4212. |
| [35] |
Liang YM, Liu HJ, Yan JB, Tian F (2021). Natural variation in crops: realized understanding, continuing promise. Annu Rev Plant Biol 72, 357-385.
DOI PMID |
| [36] |
Liu L, Song W, Huang SJ, Jiang K, Moriwaki Y, Wang YC, Men YF, Zhang D, Wen X, Han ZF, Chai JJ, Guo HW (2022a). Extracellular pH sensing by plant cell-surface peptide-receptor complexes. Cell 185, 3341-3355.
DOI URL |
| [37] |
Liu Y, Wang SB, Li LZ, Yang T, Dong SS, Wei T, Wu SD, Liu YB, Gong YQ, Feng XY, Ma JC, Chang GX, Huang JL, Yang Y, Wang HL, Liu M, Xu Y, Liang HP, Yu J, Cai YQ, Zhang ZW, Fan YN, Mu WX, Sahu SK, Liu SC, Lang XA, Yang LL, Li N, Habib S, Yang YQ, Lindstrom AJ, Liang P, Goffinet B, Zaman S, Wegrzyn JL, Li DX, Liu J, Cui J, Sonnenschein EC, Wang XB, Ruan J, Xue JY, Shao ZQ, Song C, Fan GY, Li Z, Zhang LS, Liu JQ, Liu ZJ, Jiao YN, Wang XQ, Wu H, Wang ET, Lisby M, Yang HM, Wang J, Liu X, Xu X, Li N, Soltis PS, Van de Peer Y, Soltis DE, Gong X, Liu H, Zhang SZ (2022b). The Cycas genome and the early evolution of seed plants. Nat Plants 8, 389-401.
DOI |
| [38] |
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang LJ, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao ZM, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng PZ, Bidney D, Falco C, Register J, Zhao ZY, Xu DP, Jones T, Gordon-Kamm W (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998-2015.
DOI URL |
| [39] | Pandey BK, Huang GQ, Bhosale R, Hartman S, Sturrock CJ, Jose L, Martin OC, Karady M, Voesenek LACJ, Ljung K, Lynch JP, Brown KM, Whalley WR, Mooney SJ, Zhang DB, Bennett MJ (2021). Plant roots sense soil compaction through restricted ethylene diffusion. Science 371, 276-280. |
| [40] |
Pavan S, Jacobsen E, Visser RGF, Bai YL (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed 25, 1-12.
DOI URL |
| [41] |
Schulze-Lefert P, Vogel J (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci 5, 343-348.
PMID |
| [42] |
Shang LG, Li XX, He HY, Yuan QL, Song YN, Wei ZR, Lin H, Hu M, Zhao FL, Zhang C, Li YH, Gao HS, Wang TY, Liu XP, Zhang H, Zhang Y, Cao SM, Yu XM, Zhang BT, Zhang Y, Tan YQ, Qin M, Ai C, Yang YX, Zhang B, Hu ZQ, Wang HR, Lv Y, Wang YX, Ma J, Wang Q, Lu HW, Wu Z, Liu SL, Sun ZY, Zhang HL, Guo LB, Li ZC, Zhou YF, Li JY, Zhu ZF, Xiong GS, Ruan J, Qian Q (2022). A super pan-genomic landscape of rice. Cell Res 32, 878-896.
DOI PMID |
| [43] |
Stull GW, Qu XJ, Parins-Fukuchi C, Yang YY, Yang JB, Yang ZY, Hu Y, Ma H, Soltis PS, Soltis DE, Li DZ, Smith SA, Yi TS (2021). Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nat Plants 7, 1015-1025.
DOI PMID |
| [44] | Su NN, Zhu AQ, Tao X, Ding ZJ, Chang SH, Ye F, Zhang Y, Zhao C, Chen Q, Wang JQ, Zhou CY, Guo YR, Jiao SS, Zhang SF, Wen H, Ma LX, Ye S, Zheng SJ, Yang F, Wu S, Guo JT (2022). Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature 609, 616-621. |
| [45] |
Sun Y, Wang Y, Zhang XX, Chen ZD, Xia YQ, Wang L, Sun YJ, Zhang MM, Xiao Y, Han ZF, Wang YC, Chai JJ (2022). Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature 610, 335-342.
DOI |
| [46] |
Suzuki Y, Makino A (2012). Availability of rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol 160, 533-540.
DOI PMID |
| [47] |
Tang D, Jia YX, Zhang JZ, Li HB, Cheng L, Wang P, Bao ZG, Liu ZH, Feng SS, Zhu XJ, Li DW, Zhu GT, Wang HR, Zhou Y, Zhou YF, Bryan GJ, Buell CR, Zhang CZ, Huang SW (2022). Genome evolution and diversity of wild and cultivated potatoes. Nature 606, 535-541.
DOI |
| [48] |
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189-195.
DOI |
| [49] |
Tsai HH, Schmidt W (2021). The enigma of environmental pH sensing in plants. Nat Plants 7, 106-115.
DOI |
| [50] |
Wan WL, Fröhlich K, Pruitt RN, Nürnberger T, Zhang LS (2019). Plant cell surface immune receptor complex signaling. Curr Opin Plant Biol 50, 18-28.
DOI URL |
| [51] |
Wang K, Shi L, Liang XN, Zhao P, Wang WX, Liu JX, Chang YN, Hiei Y, Yanagihara C, Du LP, Ishida Y, Ye XG (2022a). The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat Plants 8, 110-117.
DOI |
| [52] |
Wang N, Tang CL, Fan X, He MY, Gan PF, Zhang S, Hu ZY, Wang XD, Yan T, Shu WX, Yu LG, Zhao JR, He JN, Li LL, Wang JF, Huang XL, Huang LL, Zhou JM, Kang ZS, Wang XJ (2022b). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961-2974.
DOI URL |
| [53] |
Wang YP, Cheng X, Shan QW, Zhang Y, Liu JX, Gao CX, Qiu JL (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32, 947-951.
DOI PMID |
| [54] | Wei SB, Li X, Lu ZF, Zhang H, Ye XY, Zhou YJ, Li J, Yan YY, Pei HC, Duan FY, Wang DY, Chen S, Wang P, Zhang C, Shang LG, Zhou Y, Yan P, Zhao M, Huang JR, Bock R, Qian Q, Zhou WB (2022). A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 377, eabi8455. |
| [55] |
Xu F, Tang JY, Wang SX, Cheng X, Wang HR, Ou SJ, Gao SP, Li BS, Qian YW, Gao CX, Chu CC (2022). Antagonistic control of seed dormancy in rice by two bHLH transcription factors. Nat Genet 54, 1972-1982.
DOI PMID |
| [56] | Yang ZS, Xia J, Hong JJ, Zhang CX, Wei H, Ying W, Sun CQ, Sun LHX, Mao YB, Gao YX, Tan ST, Friml J, Li DF, Liu X, Sun LF (2022). Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature 609, 611-615. |
| [57] |
Yu S, Lian H, Wang JW (2015). Plant developmental transitions: the role of microRNAs and sugars. Curr Opin Plant Biol 27, 1-7.
DOI PMID |
| [58] |
Zhang H, Zhou JF, Kan Y, Shan JX, Ye WW, Dong NQ, Guo T, Xiang YH, Yang YB, Li YC, Zhao HY, Yu HX, Lu ZQ, Guo SQ, Lei JJ, Liao B, Mu XR, Cao YJ, Yu JJ, Lin YS, Lin HX (2022a). A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376, 1293-1300.
DOI URL |
| [59] |
Zhang J, Huang QP, Zhong S, Bleckmann A, Huang JY, Guo XY, Lin Q, Gu HY, Dong J, Dresselhaus T, Qu LJ (2017). Sperm cells are passive cargo of the pollen tube in plant fertilization. Nat Plants 3, 17079.
DOI PMID |
| [60] |
Zhang JS, Zhang XT, Tang HB, Zhang Q, Hua XT, Ma XK, Zhu F, Jones T, Zhu XG, Bowers J, Wai CM, Zheng CF, Shi Y, Chen S, Xu XM, Yue JJ, Nelson DR, Huang LX, Li Z, Xu HM, Zhou D, Wang YJ, Hu WC, Lin JS, Deng YJ, Pandey N, Mancini M, Zerpa D, Nguyen JK, Wang LM, Yu L, Xin YH, Ge LF, Arro J, Han JO, Chakrabarty S, Pushko M, Zhang WP, Ma YH, Ma PP, Lv MJ, Chen FM, Zheng GY, Xu JS, Yang ZH, Deng F, Chen XQ, Liao ZY, Zhang XX, Lin ZC, Lin H, Yan HS, Kuang Z, Zhong WM, Liang PP, Wang GF, Yuan Y, Shi JX, Hou JX, Lin JX, Jin JJ, Cao PJ, Shen QC, Jiang Q, Zhou P, Ma YY, Zhang XD, Xu RR, Liu J, Zhou YM, Jia HF, Ma Q, Qi R, Zhang ZL, Fang JP, Fang HK, Song JJ, Wang MJ, Dong GR, Wang G, Chen Z, Ma T, Liu H, Dhungana SR, Huss SE, Yang XP, Sharma A, Trujillo JH, Martinez MC, Hudson M, Riascos JJ, Schuler M, Chen LQ, Braun DM, Li L, Yu QY, Wang JP, Wang K, Schatz MC, Heckerman D, Van Sluys MA, Souza GM, Moore PH, Sankoff D, VanBuren R, Paterson AH, Nagai C, Ming R (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50, 1565-1573.
DOI |
| [61] |
Zhang JY, Liu YX, Zhang N, Hu B, Jin T, Xu HR, Qin Y, Yan PX, Zhang XN, Guo XX, Hui J, Cao SY, Wang X, Wang C, Wang H, Qu BY, Fan GY, Yuan LX, Garrido-Oter R, Chu CC, Bai Y (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37, 676-684.
DOI |
| [62] | Zhang Q, Qi YY, Pan HR, Tang HB, Wang G, Hua XT, Wang YJ, Lin LY, Li Z, Li YH, Yu F, Yu ZH, Huang YJ, Wang TY, Ma PP, Dou MJ, Sun ZY, Wang YB, Wang HB, Zhang XT, Yao W, Wang YT, Liu XL, Wang MJ, Wang JP, Deng ZH, Xu JS, Yang QH, Liu ZJ, Chen BS, Zhang MQ, Ming R, Zhang JS (2022b). Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat Genet 54, 885-896. |
| [63] |
Zhao H, Zhang Y, Zhang H, Song YZ, Zhao F, Zhang YE, Zhu SH, Zhang HK, Zhou ZD, Guo H, Li MM, Li JH, Gao Q, Han QQ, Huang HQ, Copsey L, Li Q, Chen H, Coen E, Zhang YJ, Xue YB (2022). Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell 34, 579-596.
DOI URL |
| [64] |
Zhao Q, Feng Q, Lu HY, Li Y, Wang AH, Tian QL, Zhan QL, Lu YQ, Zhang L, Huang T, Wang YC, Fan DL, Zhao Y, Wang ZQ, Zhou CC, Chen JY, Zhu CR, Li WJ, Weng QJ, Xu Q, Wang ZX, Wei XH, Han B, Huang XH (2018). Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50, 278-284.
DOI PMID |
| [65] | Zhong S, Li L, Wang ZJ, Ge ZX, Li QY, Bleckmann A, Wang JZ, Song ZH, Shi YH, Liu TX, Li LH, Zhou HB, Wang YY, Zhang L, Wu HM, Lai LH, Gu HY, Dong J, Cheung AY, Dresselhaus T, Qu LJ (2022). RALF peptide signaling controls the polytubey block in Arabidopsis. Science 375, 290-296. |
| [66] | Zhong S, Liu ML, Wang ZJ, Huang QP, Hou SY, Xu YC, Ge ZX, Song ZH, Huang JY, Qiu XY, Shi YH, Xiao JY, Liu P, Guo YL, Dong J, Dresselhaus T, Gu HY, Qu LJ (2019). Cysteine-rich peptides promote interspecific genetic isolation in Arabidopsis. Science 364, eaau9564. |
| [67] |
Zhou Q, Tang D, Huang W, Yang ZM, Zhang Y, Hamilton JP, Visser RGF, Bachem CWB, Robin Buell C, Zhang ZH, Zhang CZ, Huang SW (2020). Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat Genet 52, 1018-1023.
DOI PMID |
| [68] |
Zhou Y, Zhang ZY, Bao ZG, Li HB, Lyu YQ, Zan YJ, Wu YY, Cheng L, Fang YH, Wu K, Zhang JZ, Lyu HJ, Lin T, Gao Q, Saha S, Mueller L, Fei ZJ, Städler T, Xu SZ, Zhang ZW, Speed D, Huang SW (2022). Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527-534.
DOI |
| [1] | 韩鹏宾 裴康迪 何斌 肖书礼 唐勤. 中国云南铁杉林的群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [2] | 何斌 韩鹏宾 肖书礼 裴康迪 唐勤. 铁坚油杉林的群落类型及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 魏梦璐, 李锦粤, 饶玉春. 基于“以赛促创”的植物科学领域创新型人才培养实践[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 田志奇, 苏杨. 环境相关国际公约的中国履约模式和在《生物多样性公约》中的应用: 从完成《昆蒙框架》目标和发挥国家公园作用的角度[J]. 生物多样性, 2025, 33(3): 24593-. |
| [5] | 周志华, 金效华, 罗颖, 李迪强, 岳建兵, 刘芳, 何拓, 李希, 董晖, 罗鹏. 中国林草部门落实《昆明-蒙特利尔全球生物多样性框架》的机制、成效分析及建议[J]. 生物多样性, 2025, 33(3): 24487-. |
| [6] | 顾红雅, 陈凡, 林荣呈, 漆小泉, 杨淑华, 陈之端, 陈学伟, 丁兆军, 萧浪涛, 左建儒, 姜里文, 白永飞, 种康, 王雷. 2024年中国植物科学重要研究进展[J]. 植物学报, 2025, 60(2): 151-171. |
| [7] | 宋阳, 柳军, 何少林, 徐薇, 程琛, 刘博, 余绩庆. 我国能源企业生物多样性保护主流化管理路径[J]. 生物多样性, 2025, 33(1): 24345-. |
| [8] | 弋维, 艾鷖, 吴萌, 田黎明, 泽让东科. 青藏高原高寒草甸土壤古菌群落对不同放牧强度的响应[J]. 生物多样性, 2025, 33(1): 24339-. |
| [9] | 耿江天, 王菲, 赵华斌. 城市化对中国蝙蝠影响的研究进展[J]. 生物多样性, 2024, 32(8): 24109-. |
| [10] | 李邦泽, 张树仁. 中国莎草科最新物种名录和分类纲要[J]. 生物多样性, 2024, 32(7): 24106-. |
| [11] | 胡宗刚. 抗战胜利后中美曾筹划合编《中国植物志》[J]. 生物多样性, 2024, 32(6): 24220-. |
| [12] | 池玉杰, 张心甜, 田志炫, 关成帅, 谷新治, 刘智会, 王占斌, 王金杰. 东北亚地区白粉菌的物种多样性与寄主物种多样性[J]. 生物多样性, 2024, 32(4): 23443-. |
| [13] | 盘远方, 潘良浩, 邱思婷, 邱广龙, 苏治南, 史小芳, 范航清. 中国沿海红树林树高变异与环境适应机制[J]. 植物生态学报, 2024, 48(4): 483-495. |
| [14] | 刘荆州, 钱易鑫, 张燕雪丹, 崔凤. 基于潜在迪利克雷分布(LDA)模型的旗舰物种范式研究进展与启示[J]. 生物多样性, 2024, 32(4): 23439-. |
| [15] | 陈凡, 顾红雅, 漆小泉, 林荣呈, 钱前, 萧浪涛, 杨淑华, 左建儒, 白永飞, 陈之端, 丁兆军, 王小菁, 姜里文, 种康, 王雷. 2023年中国植物科学重要研究进展[J]. 植物学报, 2024, 59(2): 171-187. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||