

植物学报 ›› 2023, Vol. 58 ›› Issue (2): 316-334.DOI: 10.11983/CBB22030 cstr: 32102.14.CBB22030
所属专题: 大食物观
郭彦君1, 陈枫1, 罗敬文1, 曾为2, 许文亮1,*( )
)
收稿日期:2022-02-21
接受日期:2022-06-23
出版日期:2023-03-01
发布日期:2023-03-15
通讯作者:
*E-mail: 基金资助:
Yanjun Guo1, Feng Chen1, Jingwen Luo1, Wei Zeng2, Wenliang Xu1,*( )
)
Received:2022-02-21
Accepted:2022-06-23
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: 摘要: 木聚糖是广泛存在于各类植物细胞壁中的半纤维素, 对植物生长发育至关重要。许多研究表明, 细胞壁中木聚糖的含量和结构对生物质的加工特性有显著影响。因此, 理解木聚糖的生物合成机制有助于利用基因工程手段对细胞壁进行改良。近10年来, 在模式植物拟南芥(Arabidopsis thaliana)以及重要粮食和经济作物中鉴定出许多参与木聚糖生物合成的基因。该文综述了相关研究进展, 并探讨了木聚糖生物合成基因在生物质能及相关领域的潜在应用价值。
郭彦君, 陈枫, 罗敬文, 曾为, 许文亮. 植物细胞壁木聚糖的生物合成及其应用. 植物学报, 2023, 58(2): 316-334.
Yanjun Guo, Feng Chen, Jingwen Luo, Wei Zeng, Wenliang Xu. The Biosynthesis of Plant Cell Wall Xylan and Its Application. Chinese Bulletin of Botany, 2023, 58(2): 316-334.
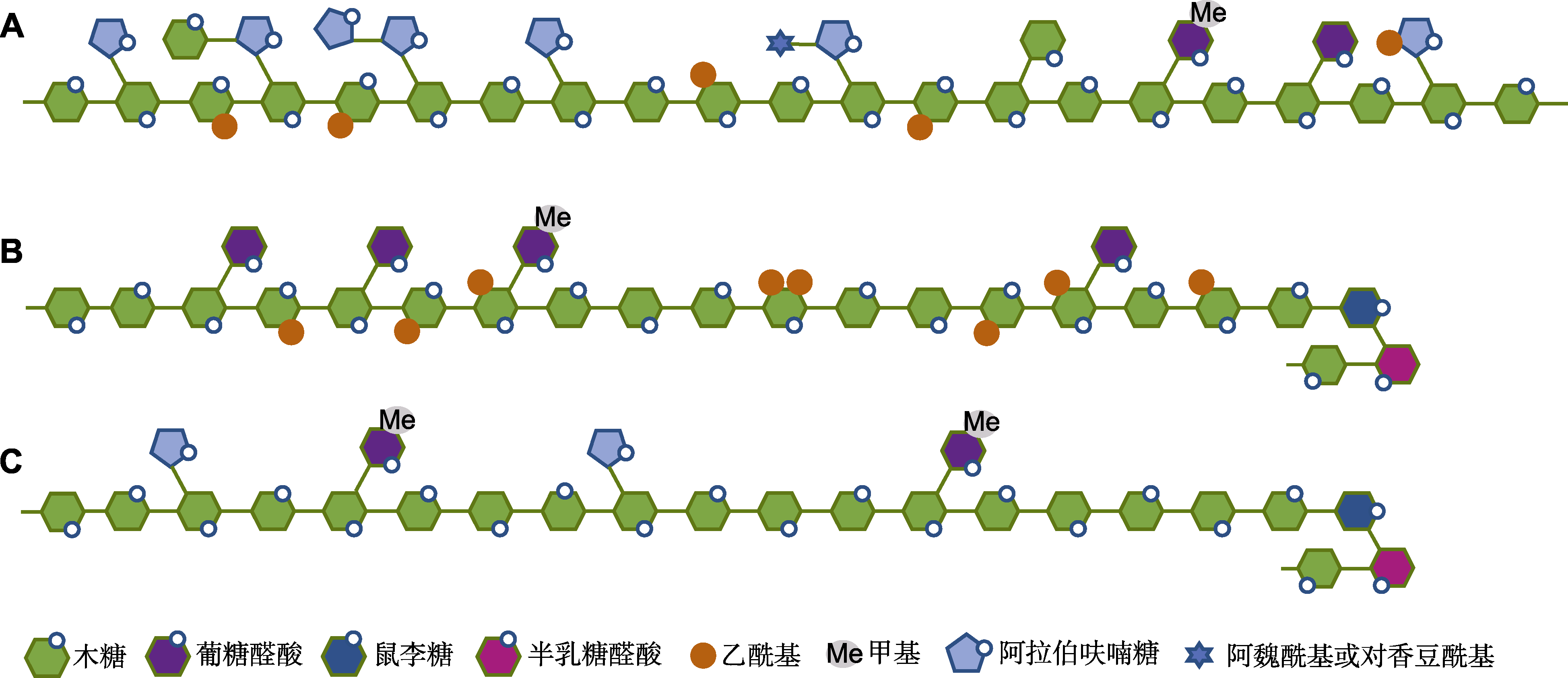
图1 木聚糖结构示意图(参考Zhong et al., 2019; Zhang et al., 2021) (A) 单子叶禾本科植物中的葡糖醛酸阿拉伯木聚糖; (B) 双子叶植物及一些非禾本科单子叶植物中的葡糖醛酸木聚糖; (C) 裸子植物中的葡糖醛酸阿拉伯木聚糖
Figure 1 Generalized structures of xylan (refer to Zhong et al., 2019; Zhang et al., 2021) (A) Glucuronoarabinoxylan in the plants of Poaceae; (B) Glucuronoxylan in dicot and some non-Poaceae monocotyledonous plants; (C) Glucuronoarabinoxylan in gymnosperm
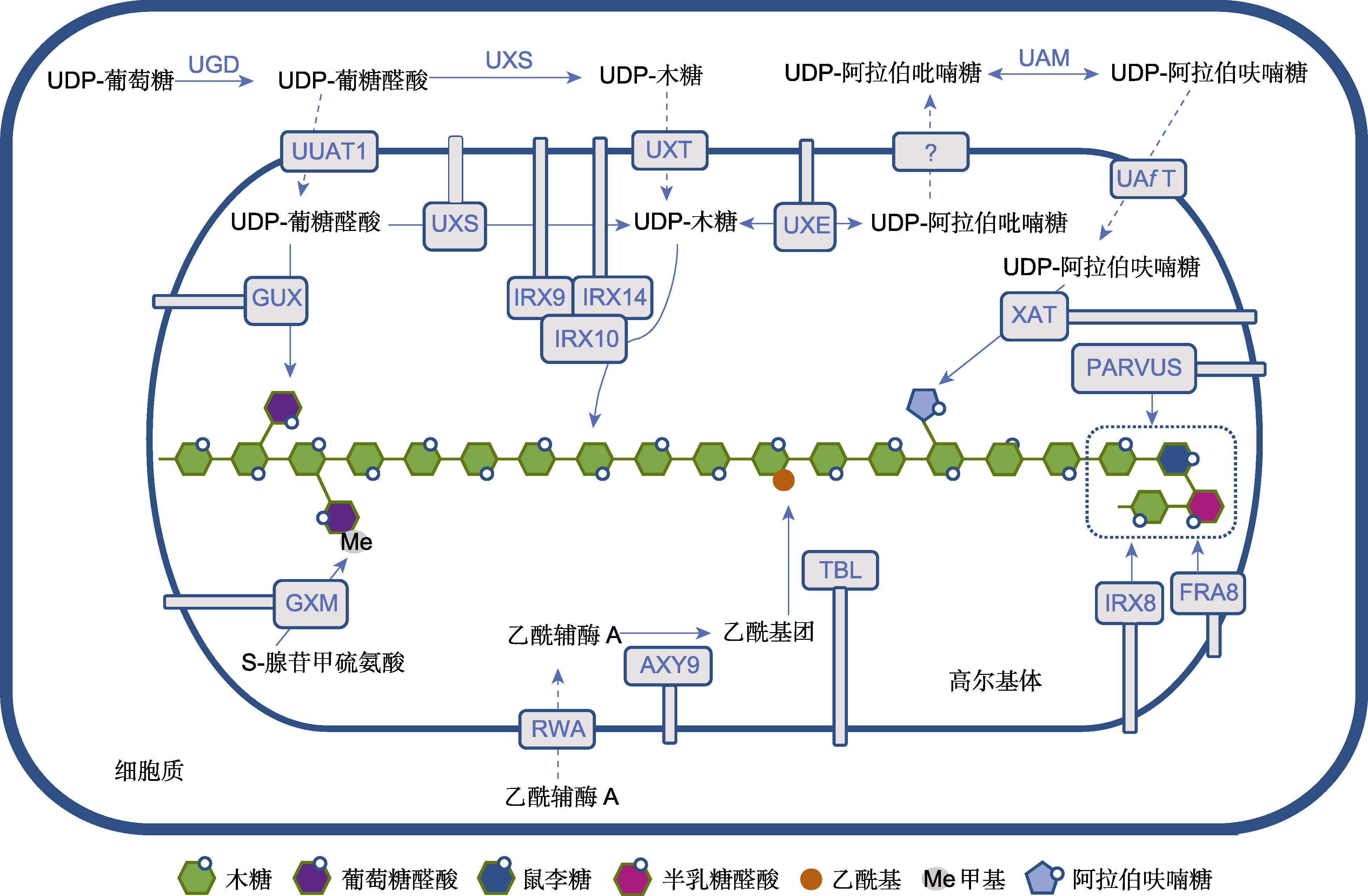
图2 木聚糖生物合成示意图(参考Qaseem and Wu, 2020) UGD: UDP-葡萄糖脱氢酶; UXS: UDP-木糖合成酶; UAM: UDP-阿拉伯糖变位酶; UUAT1: UDP-糖醛酸转运蛋白; UXT: UDP-木糖转运蛋白; UAfT: UDP-阿拉伯糖转运蛋白; UXE: UDP-木糖差向异构酶。虚线箭头表示转运过程, 实线箭头表示化学反应过程, 问号表示未知。
Figure 2 Schematic diagram of xylan biosynthesis (refer to Qaseem and Wu, 2020) UGD: UDP-Glc dehydrogenase; UXS: UDP-Xyl synthase; UAM: UDP-Ara mutase; UUAT1: UDP-Uronic acid transporter 1; UXT: UDP-Xyl transporter; UAfT: UDP-Araf transporter; UXE: UDP-Xyl epimerase. Dotted arrows show the transport process, the solid arrows show the chemical reaction process, question mark indicates unknown.
| 物种 | 基因 | 功能 | 应用 | 参考文献 |
|---|---|---|---|---|
| 二穗短柄草(Brachypodium distachyon) | BdGT43A/B2 | 参与主链合成 | 提高生物质消化率 | Whitehead et al., |
| BdBAHD01/05 | 参与羟基肉桂酸取代 | ND | Buanafina et al., | |
| 小麦(Triticum aestivum) | TaGT43_1/2, TaGT47_2 | 参与主链合成 | 获得更易提取的小麦籽粒水提物 | Jiang et al., |
| TaXAT1/2 | 阿拉伯糖基转移酶 | 获得更易提取的小麦籽粒水提物 | Anders et al., | |
| 水稻(Oryza sativa) | OsGT43A/B/E/J | 参与主链合成 | ND | Lee et al., |
| OsIRX10 | 参与主链合成 | 提高生物质消化率 | Chen et al., | |
| OsGUX1 | 葡糖醛酸转移酶 | 改变叶绿素含量 | Gao et al., | |
| OsXAT2/3/4/5/6/7 | 阿拉伯糖基转移酶 | ND | Zhong et al., | |
| OsXAX1 | 不明确 | 提高生物质消化率 | Chiniquy et al., | |
| OsXYXT1 | 向主链添加木糖侧链 | ND | Zhong et al., | |
| OsTBL1/2 | 参与主链乙酰化 | 增强抗病性 | Gao et al., | |
| OsXOAT1-14 | 参与主链乙酰化 | ND | Zhong et al., | |
| OsBS1 | 参与主链去乙酰化 | 提高机械强度 | Zhang et al., | |
| OsDARX1 | 参与阿拉伯糖残基侧链去乙酰化 | 提高机械强度 | Zhang et al., | |
| OsAT10 | 参与羟基肉桂酸取代 | 提高生物质消化率 | Bartley et al., | |
| OsFC18 | 合成UDP-木糖 | 提高生物质消化率 | Ruan et al., | |
| 狗尾草(Setaria viridis) | SvBAHD01/05 | 参与羟基肉桂酸取代 | 提高生物质消化率 | de Souza et al., |
| 甘蔗(Saccharum officinarum) | SacBAHD01 | 参与羟基肉桂酸取代 | 提高生物质消化率 | de Souza et al., |
| 杨树(Populus spp.) | PtrGT43A/B/C/D/E/F/G | 参与主链合成 | 提高生物质消化率 | Lee et al., |
| PoGT47C | 参与还原末端合成 | 提高生物质消化率 | Lee et al., | |
| PoGT8D/E/F | 参与还原末端合成 | ND | Lee et al., | |
| Pt/PdGAUT12 | 参与还原末端合成 | 提高生物质消化率 | Biswal et al., | |
| PtrGXM1-4 | 甲基转移酶 | 提高生物质消化率 | Song et al., | |
| PtrXOAT1-12 | 乙酰基转移酶 | ND | Zhong et al., | |
| 云杉(Picea glauca) | PgGUX | 葡糖醛酸转移酶 | ND | Lyczakowski et al., |
| 陆地棉(Gossypium hirsutum) | GhGT43A1/C1 | 参与主链合成 | ND | Li et al., |
| GhGT47A1 | 参与主链合成 | ND | Chen et al., | |
| GhGT47B1/B2 | 参与还原末端合成 | ND | Chen et al., |
表1 不同物种木聚糖生物合成基因及应用
Table 1 Xylan biosynthesis genes in different species and their applications
| 物种 | 基因 | 功能 | 应用 | 参考文献 |
|---|---|---|---|---|
| 二穗短柄草(Brachypodium distachyon) | BdGT43A/B2 | 参与主链合成 | 提高生物质消化率 | Whitehead et al., |
| BdBAHD01/05 | 参与羟基肉桂酸取代 | ND | Buanafina et al., | |
| 小麦(Triticum aestivum) | TaGT43_1/2, TaGT47_2 | 参与主链合成 | 获得更易提取的小麦籽粒水提物 | Jiang et al., |
| TaXAT1/2 | 阿拉伯糖基转移酶 | 获得更易提取的小麦籽粒水提物 | Anders et al., | |
| 水稻(Oryza sativa) | OsGT43A/B/E/J | 参与主链合成 | ND | Lee et al., |
| OsIRX10 | 参与主链合成 | 提高生物质消化率 | Chen et al., | |
| OsGUX1 | 葡糖醛酸转移酶 | 改变叶绿素含量 | Gao et al., | |
| OsXAT2/3/4/5/6/7 | 阿拉伯糖基转移酶 | ND | Zhong et al., | |
| OsXAX1 | 不明确 | 提高生物质消化率 | Chiniquy et al., | |
| OsXYXT1 | 向主链添加木糖侧链 | ND | Zhong et al., | |
| OsTBL1/2 | 参与主链乙酰化 | 增强抗病性 | Gao et al., | |
| OsXOAT1-14 | 参与主链乙酰化 | ND | Zhong et al., | |
| OsBS1 | 参与主链去乙酰化 | 提高机械强度 | Zhang et al., | |
| OsDARX1 | 参与阿拉伯糖残基侧链去乙酰化 | 提高机械强度 | Zhang et al., | |
| OsAT10 | 参与羟基肉桂酸取代 | 提高生物质消化率 | Bartley et al., | |
| OsFC18 | 合成UDP-木糖 | 提高生物质消化率 | Ruan et al., | |
| 狗尾草(Setaria viridis) | SvBAHD01/05 | 参与羟基肉桂酸取代 | 提高生物质消化率 | de Souza et al., |
| 甘蔗(Saccharum officinarum) | SacBAHD01 | 参与羟基肉桂酸取代 | 提高生物质消化率 | de Souza et al., |
| 杨树(Populus spp.) | PtrGT43A/B/C/D/E/F/G | 参与主链合成 | 提高生物质消化率 | Lee et al., |
| PoGT47C | 参与还原末端合成 | 提高生物质消化率 | Lee et al., | |
| PoGT8D/E/F | 参与还原末端合成 | ND | Lee et al., | |
| Pt/PdGAUT12 | 参与还原末端合成 | 提高生物质消化率 | Biswal et al., | |
| PtrGXM1-4 | 甲基转移酶 | 提高生物质消化率 | Song et al., | |
| PtrXOAT1-12 | 乙酰基转移酶 | ND | Zhong et al., | |
| 云杉(Picea glauca) | PgGUX | 葡糖醛酸转移酶 | ND | Lyczakowski et al., |
| 陆地棉(Gossypium hirsutum) | GhGT43A1/C1 | 参与主链合成 | ND | Li et al., |
| GhGT47A1 | 参与主链合成 | ND | Chen et al., | |
| GhGT47B1/B2 | 参与还原末端合成 | ND | Chen et al., |
| [1] | 刘妍, 邱爽, 陈浩, 庞晓晗, 辰巳英三, 殷丽君 (2015). 玉米纤维胶的制备和特性研究进展. 中国食物与营养 21(6), 33-37. |
| [2] | 秦丽霞, 张德静, 李龙, 李学宝, 许文亮 (2011). 参与植物细胞壁半纤维素木聚糖合成的糖基转移酶. 植物生理学报 47, 831-839. |
| [3] | 宋晓庆, 董海洲 (2004). 黑麦阿拉伯木聚糖的结构、性质与营养功能. 中国食物与营养 (2), 28-29, 58. |
| [4] |
张雨, 赵明洁, 张蔚 (2020). 植物次生细胞壁生物合成的转录调控网络. 植物学报 55, 351-368.
DOI |
| [5] |
Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, Defoin-Platel M, Shewry PR, Dupree P, Mitchell RA (2012). Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA 109, 989-993.
DOI PMID |
| [6] |
Bar-Peled M, O’Neill MA (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu Rev Plant Biol 62, 127-155.
DOI PMID |
| [7] |
Bartley LE, Peck ML, Kim SR, Ebert B, Manisseri C, Chiniquy DM, Sykes R, Gao LF, Rautengarten C, Vega- Sánchez ME, Benke PI, Canlas PE, Cao PJ, Brewer S, Lin F, Smith WL, Zhang XH, Keasling JD, Jentoff RE, Foster SB, Zhou JZ, Ziebell A, An G, Scheller HV, Ronald PC (2013). Overexpression of a BAHD acyltransferase, OsAT10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol 161, 1615-1633.
DOI PMID |
| [8] | Biswal AK, Atmodjo MA, Pattathil S, Amos RA, Yang XH, Winkeler K, Collins C, Mohanty SS, Ryno D, Tan L, Gelineo-Albersheim I, Hunt K, Sykes RW, Turner GB, Ziebell A, Davis MF, Decker SR, Hahn MG, Mohnen D (2018). Working towards recalcitrance mechanisms: increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12 (GAUT12) causes increased recalcitrance and decreased growth in Populus. Biotechnol Biofuels 11, 9. |
| [9] | Biswal AK, Hao ZY, Pattathil S, Yang XH, Winkeler K, Collins C, Mohanty SS, Richardson EA, Gelineo-Albersheim I, Hunt K, Ryno D, Sykes RW, Turner GB, Ziebell A, Gjersing E, Lukowitz W, Davis MF, Decker SR, Hahn MG, Mohnen D (2015). Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol Biofuels 8, 41. |
| [10] |
Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang ZN, Brown DM, Dupree P (2013). GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J 74, 423-434.
DOI URL |
| [11] |
Brown D, Wightman R, Zhang ZN, Gomez LD, Atanassov I, Bukowski JP, Tryfona T, McQueen-Mason SJ, Dupree P, Turner S (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J 66, 401-413.
DOI URL |
| [12] |
Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR (2007). Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52, 1154-1168.
DOI PMID |
| [13] |
Brown DM, Zhang ZN, Stephens E, Dupree P, Turner SR (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57, 732-746.
DOI URL |
| [14] |
Broxterman SE, Schols HA (2018). Characterisation of pectin-xylan complexes in tomato primary plant cell walls. Carbohydr Polym 197, 269-276.
DOI URL |
| [15] |
Bryant ND, Pu YQ, Tschaplinski TJ, Tuskan GA, Muchero W, Kalluri UC, Yoo CG, Ragauskas AJ (2020). Transgenic poplar designed for biofuels. Trends Plant Sci 25, 881-896.
DOI PMID |
| [16] |
Buanafina MMD, Fescemyer HW, Sharma M, Shearer EA (2016). Functional testing of a PF02458 homologue of putative rice arabinoxylan feruloyl transferase genes in Brachypodium distachyon. Planta 243, 659-674.
DOI PMID |
| [17] |
Burget EG, Verma R, Mølhøj M, Reiter WD (2003). The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15, 523-531.
DOI URL |
| [18] |
Busse-Wicher M, Li A, Silveira RL, Pereira CS, Tryfona T, Gomes TCF, Skaf MS, Dupree P (2016). Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiol 171, 2418-2431.
DOI PMID |
| [19] |
Chen F, Guo YJ, Chen L, Gan XL, Liu M, Li J, Xu WL (2020). Global identification of genes associated with xylan biosynthesis in cotton fiber. J Cotton Res 3, 25.
DOI |
| [20] |
Chen XW, Vega-Sánchez ME, Verhertbruggen Y, Chiniquy D, Canlas PE, Fagerström A, Prak L, Christensen U, Oikawa A, Chern M, Zuo SM, Lin F, Auer M, Willats WGT, Bartley L, Harholt J, Scheller HV, Ronald PC (2013). Inactivation of OsIRX10 leads to decreased xylan content in rice culm cell walls and improved biomass saccharification. Mol Plant 6, 570-573.
DOI URL |
| [21] |
Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, Pauly M, Scheller HV, Ronald PC (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA 109, 17117-17122.
DOI PMID |
| [22] | Crowe JD, Hao PC, Pattathil S, Pan H, Ding SY, Hodge DB, Jensen JK (2021). Xylan is critical for proper bundling and alignment of cellulose microfibrils in plant secondary cell walls. Front Plant Sci 12, 737690. |
| [23] |
De Carvalho DM, Marchand C, Berglund J, Lindström ME, Vilaplana F, Sevastyanova O (2020). Impact of birch xylan composition and structure on film formation and properties. Holzforschung 74, 184-196.
DOI URL |
| [24] |
De Souza WR, Martins PK, Freeman J, Pellny TK, Michaelson LV, Sampaio BL, Vinecky F, Ribeiro AP, Da Cunha BADB, Kobayashi AK, De Oliveira PA, Campanha RB, Pacheco TF, Martarello DCI, Marchiosi R, Ferrarese-Filho O, Dos Santos WD, Tramontina R, Squina FM, Centeno DC, Gaspar M, Braga MR, Tiné MAS, Ralph J, Mitchell RAC, Molinari HBC (2018). Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytol 218, 81-93.
DOI URL |
| [25] |
De Souza WR, Pacheco TF, Duarte KE, Sampaio BL, De Oliveira Molinari PA, Martins PK, Santiago TR, Formighieri EF, Vinecky F, Ribeiro AP, Da Cunha BADB, Kobayashi AK, Mitchell RAC, De Sousa Rodrigues Gambetta D, Molinari HBC (2019). Silencing of a BAHD acyltransferase in sugarcane increases biomass digestibility. Biotechnol Biofuels 12, 111.
DOI PMID |
| [26] |
Deniaud E, Quemener B, Fleurence J, Lahaye M (2003). Structural studies of the mix-linked β(1→3)/β(1→4)-D-xylans from the cell wall of Palmaria palmata (Rhodophyta). Int J Biol Macromol 33, 9-18.
DOI URL |
| [27] |
Feijao C, Morreel K, Anders N, Tryfona T, Busse-Wicher M, Kotake T, Boerjan W, Dupree P (2022). Hydroxycinnamic acid-modified xylan side chains and their cross-linking products in rice cell walls are reduced in the Xylosyl arabinosyl substitution of xylan 1mutant. Plant J 109, 1152-1167.
DOI URL |
| [28] |
Freeman J, Lovegrove A, Wilkinson MD, Saulnier L, Shewry PR, Mitchell RAC (2016). Effect of suppression of arabinoxylan synthetic genes in wheat endosperm on chain length of arabinoxylan and extract viscosity. Plant Biotechnol J 14, 109-116.
DOI PMID |
| [29] |
Gao DW, Sun WQ, Wang DW, Dong HL, Zhang R, Yu SB (2020a). A xylan glucuronosyltransferase gene exhibits pleiotropic effects on cellular composition and leaf development in rice. Sci Rep 10, 3726.
DOI |
| [30] |
Gao Y, Lipton AS, Wittmer Y, Murray DT, Mortimer JC (2020b). A grass-specific cellulose-xylan interaction dominates in sorghum secondary cell walls. Nat Commun 11, 6081.
DOI |
| [31] |
Gao YP, He CW, Zhang DM, Liu XL, Xu ZP, Tian YB, Liu XH, Zang SS, Pauly M, Zhou YH, Zhang BC (2017). Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol 173, 470-481.
DOI PMID |
| [32] |
Gille S, Pauly M (2012). O-acetylation of plant cell wall polysaccharides. Front Plant Sci 3, 12.
DOI PMID |
| [33] |
Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, Busse-Wicher M, Dupree P (2017). An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants 3, 859-865.
DOI PMID |
| [34] | Haghighat M, Teng Q, Zhong RQ, Ye ZH (2016). Evolutionary conservation of xylan biosynthetic genes in Selaginella moellendorffii and Physcomitrella patens. Plant Cell Physiol 57, 1707-1719. |
| [35] | Hatfield RD, Rancour DM, Marita JM (2017). Grass cell walls: a story of cross-linking. Front Plant Sci 7, 2056. |
| [36] | He JB, Zhao XH, Du PZ, Zeng W, Beahan CT, Wang YQ, Li HL, Bacic A, Wu AM (2018). KNAT7 positively regulates xylan biosynthesis by directly activating IRX9 expression in Arabidopsis. J Integr Plant Biol 60, 514-528. |
| [37] |
Hörnblad E, Ulfstedt M, Ronne H, Marchant A (2013). Partial functional conservation of IRX10 homologs in Physcomitrella patens and Arabidopsis thaliana indicates an evolutionary step contributing to vascular formation in land plants. BMC Plant Biol 13, 3.
DOI PMID |
| [38] |
Hu RB, Li JL, Wang XY, Zhao X, Yang XW, Tang Q, He G, Zhou GK, Kong YZ (2016a). Xylan synthesized by irregular xylem 14 (IRX14) maintains the structure of seed coat mucilage in Arabidopsis. J Exp Bot 67, 1243-1257.
DOI URL |
| [39] |
Hu RB, Li JL, Yang XW, Zhao X, Wang XY, Tang Q, He G, Zhou GK, Kong YZ (2016b). Irregular xylem 7 (IRX7) is required for anchoring seed coat mucilage in Arabidopsis. Plant Mol Biol 92, 25-38.
DOI URL |
| [40] |
Huang JF, Chen F, Guo YJ, Gan XL, Yang MM, Zeng W, Persson S, Li J, Xu WL (2021). GhMYB7 promotes secondary wall cellulose deposition in cotton fibres by regulating GhCesA gene expression through three distinct cis-elements. New Phytol 232, 1718-1737.
DOI URL |
| [41] |
Huang JF, Guo YJ, Sun QW, Zeng W, Li J, Li XB, Xu WL (2019). Genome-wide identification of R2R3-MYB transcription factors regulating secondary cell wall thickening in cotton fiber development. Plant Cell Physiol 60, 687-701.
DOI PMID |
| [42] |
Jensen JK, Busse-Wicher M, Poulsen CP, Fangel JU, Smith PJ, Yang JY, Peña MJ, Dinesen MH, Martens HJ, Melkonian M, Wong GKS, Moremen KW, Wilkerson CG, Scheller HV, Dupree P, Ulvskov P, Urbanowicz BR, Harholt J (2018). Identification of an algal xylan synthase indicates that there is functional orthology between algal and plant cell wall biosynthesis. New Phytol 218, 1049-1060.
DOI PMID |
| [43] |
Jensen JK, Johnson N, Wilkerson CG (2013). Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front Plant Sci 4, 183.
DOI PMID |
| [44] |
Jensen JK, Johnson NR, Wilkerson CG (2014). Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J 80, 207-215.
DOI URL |
| [45] | Jensen JK, Kim H, Cocuron JC, Orler R, Ralph J, Wilkerson CG (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J 66, 387-400. |
| [46] |
Jiang N, Wiemels RE, Soya A, Whitley R, Held M, Faik A (2016). Composition, assembly, and trafficking of a wheat xylan synthase complex. Plant Physiol 170, 1999-2023.
DOI PMID |
| [47] |
Kang X, Kirui A, Widanage MCD, Mentink-Vigier F, Cosgrove DJ, Wang T (2019). Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid- state NMR. Nat Commun 10, 347.
DOI PMID |
| [48] |
Kim WC, Kim JY, Ko JH, Kang H, Han KH (2014). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol Biol 85, 589-599.
DOI URL |
| [49] | Kuang BQ, Zhao XH, Zhou C, Zeng W, Ren JL, Ebert B, Beahan CT, Deng XM, Zeng QY, Zhou GK, Doblin MS, Heazlewood JL, Bacic A, Chen XY, Wu AM (2016). Role of UDP-glucuronic acid decarboxylase in xylan biosynthesis in Arabidopsis. Mol Plant 9, 1119-1131. |
| [50] |
Kulkarni AR, Peña MJ, Avci U, Mazumder K, Urbanowicz BR, Pattathil S, Yin YB, O'Neill MA, Roberts AW, Hahn MG, Xu Y, Darvill AG, York WS (2012). The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology 22, 439-451.
DOI PMID |
| [51] |
Kumar M, Campbell L, Turner S (2016). Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67, 515-531.
DOI PMID |
| [52] |
Lee C, Teng Q, Huang WL, Zhong RQ, Ye ZH (2009a). Down-regulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol 50, 1075-1089.
DOI URL |
| [53] | Lee C, Teng Q, Huang WL, Zhong RQ, Ye ZH (2009b). The F8H glycosyltransferase is a functional paralog of FRA8 involved in glucuronoxylan biosynthesis in Arabidopsis. Plant Cell Physiol 50, 812-827. |
| [54] |
Lee C, Teng Q, Huang WL, Zhong RQ, Ye ZH (2009c). The poplar GT8E and GT8F glycosyltransferases are functional orthologs of Arabidopsis PARVUS involved in glucuronoxylan biosynthesis. Plant Cell Physiol 50, 1982-1987.
DOI URL |
| [55] |
Lee C, Teng Q, Zhong RQ, Ye ZH (2011a). The four Arabidopsis REDUCED WALL ACETYLATION genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52, 1289-1301.
DOI URL |
| [56] |
Lee C, Teng Q, Zhong RQ, Ye ZH (2011b). Molecular dissection of xylan biosynthesis during wood formation in poplar. Mol Plant 4, 730-747.
DOI URL |
| [57] |
Lee C, Teng Q, Zhong RQ, Yuan YX, Haghighat M, Ye ZH (2012). Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol 53, 1934-1949.
DOI URL |
| [58] | Lee C, Teng Q, Zhong RQ, Yuan YX, Ye ZH (2014). Functional roles of rice glycosyltransferase family GT43 in xylan biosynthesis. Plant Signal Behav 9, e27809. |
| [59] |
Lee C, Zhong RQ, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH (2007). The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48, 1659-1672.
DOI URL |
| [60] |
Li GT, Jones KC, Eudes A, Pidatala VR, Sun J, Xu F, Zhang CC, Wei T, Jain R, Birdseye D, Canlas PE, Baidoo EEK, Duong PQ, Sharma MK, Singh S, Ruan DL, Keasling JD, Mortimer JC, Loqué D, Bartley LE, Scheller HV, Ronald PC (2018). Overexpression of a rice BAHD acyltransferase gene in switchgrass (Panicum virgatum L.) enhances saccharification. BMC Biotechnol 18, 54.
DOI |
| [61] |
Li L, Huang JF, Qin LX, Huang YY, Zeng W, Rao Y, Li J, Li XB, Xu WL (2014). Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiol Plant 152, 367-379.
DOI PMID |
| [62] |
Li QZ, Min DY, Wang JPY, Peszlen I, Horvath L, Horvath B, Nishimura Y, Jameel H, Chang HM, Chiang VL (2011). Down-regulation of glycosyltransferase 8D genes in Populus trichocarpa caused reduced mechanical strength and xylan content in wood. Tree Physiol 31, 226-236.
DOI URL |
| [63] |
Lundborg M, Fontana C, Widmalm G (2011). Automatic structure determination of regular polysaccharides based solely on NMR spectroscopy. Biomacromolecules 12, 3851-3855.
DOI PMID |
| [64] | Lunin VV, Wang HT, Bharadwaj VS, Alahuhta M, Peña MJ, Yang JY, Archer-Hartmann SA, Azadi P, Himmel ME, Moremen KW, York WS, Bomble YJ, Urbanowicz BR (2020). Molecular mechanism of polysaccharide acetylation by the Arabidopsis xylan O-acetyltransferase XOA T1. Plant Cell 32, 2367-2382. |
| [65] |
Lyczakowski JJ, Wicher KB, Terrett OM, Faria-Blanc N, Yu XL, Brown D, Krogh KBRM, Dupree P, Busse-Wicher M (2017). Removal of glucuronic acid from xylan is a strategy to improve the conversion of plant biomass to sugars for bioenergy. Biotechnol Biofuels 10, 224.
DOI PMID |
| [66] | Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong SL, Pawar PMA, Mellerowicz EJ, Tenkanen M, Cheng K, Pauly M, Scheller HV (2013). REDUCED WALL ACETYLATION proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol 163, 1107-1117. |
| [67] |
McCleary BV, McKie VA, Draga A, Rooney E, Mangan D, Larkin J (2015). Hydrolysis of wheat flour arabinoxylan, acid-debranched wheat flour arabinoxylan and arabino- xylo-oligosaccharides by β-xylanase, α-L-arabinofuranosidase and β-xylosidase. Carbohydr Res 407, 79-96.
DOI URL |
| [68] |
Meents MJ, Motani S, Mansfield SD, Samuels AL (2019). Organization of xylan production in the Golgi during secondary cell wall biosynthesis. Plant Physiol 181, 527-546.
DOI PMID |
| [69] |
Meents MJ, Watanabe Y, Samuels AL (2018). The cell biology of secondary cell wall biosynthesis. Ann Bot 121, 1107-1125.
DOI URL |
| [70] | Mota TR, De Souza WR, Oliveira DM, Martins PK, Sampaio BL, Vinecky F, Ribeiro AP, Duarte KE, Pacheco TF, Monteiro NDV, Campanha RB, Marchiosi R, Vieira DS, Kobayashi AK, Molinari PAD, Ferrarese-Filho O, Mitchell RAC, Molinari HBC, Dos Santos WD (2021). Suppression of a BAHD acyltransferase decreases p-coumaroyl on arabinoxylan and improves biomass digestibility in the model grass Setaria viridis. Plant J 105, 136-150. |
| [71] |
Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6, 288.
DOI PMID |
| [72] |
Oliveira DM, Mota TR, Salatta FV, Marchiosi R, Gomez LD, McQueen-Mason SJ, Ferrarese-Filho O, Dos Santos WD (2019). Designing xylan for improved sustainable biofuel production. Plant Biotechnol J 17, 2225-2227.
DOI PMID |
| [73] |
Pawar PMA, Derba-Maceluch M, Chong SL, Gandla ML, Bashar SS, Sparrman T, Ahvenainen P, Hedenström M, Özparpucu M, Rüggeberg M, Serimaa R, Lawoko M, Tenkanen M, Jönsson LJ, Mellerowicz EJ (2017). In muro deacetylation of xylan affects lignin properties and improves saccharification of aspen wood. Biotechnol Biofuels 10, 98.
DOI URL |
| [74] | Pawar PMA, Koutaniemi S, Tenkanen M, Mellerowicz EJ (2013). Acetylation of woody lignocellulose: significance and regulation. Front Plant Sci 4, 118. |
| [75] |
Pellny TK, Lovegrove A, Freeman J, Tosi P, Love CG, Knox JP, Shewry PR, Mitchell RAC (2012). Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-seq transcriptome. Plant Physiol 158, 612-627.
DOI PMID |
| [76] |
Pellny TK, Patil A, Wood AJ, Freeman J, Halsey K, Plummer A, Kosik O, Temple H, Collins JD, Dupree P, Berry S, Shewry PR, Lovegrove A, Phillips AL, Mitchell RAC (2020). Loss of TaIRX9b gene function in wheat decreases chain length and amount of arabinoxylan in grain but increases cross-linking. Plant Biotechnol J 18, 2316-2327.
DOI URL |
| [77] |
Peña MJ, Kulkarni AR, Backe J, Boyd M, O’Neill MA, York WS (2016). Structural diversity of xylans in the cell walls of monocots. Planta 244, 589-606.
DOI PMID |
| [78] |
Peña MJ, Zhong RQ, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH (2007). Arabidopsis irregular xylem 8 and irregular xylem 9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19, 549-563.
DOI URL |
| [79] | Petrik DL, Tryfona T, Dupree P, Anderson CT (2020). functions in xylan biosynthesis and is essential for seedling survival in Brachypodium distachyon. Plant Direct 4, e00216. |
| [80] |
Phan JL, Tucker MR, Khor SF, Shirley N, Lahnstein J, Beahan C, Bacic A, Burton RA (2016). Differences in glycosyltransferase family 61 accompany variation in seed coat mucilage composition in Plantago spp. J Exp Bot 67, 6481-6495.
DOI URL |
| [81] |
Qaseem MF, Wu AM (2020). Balanced xylan acetylation is the key regulator of plant growth and development, and cell wall structure and for industrial utilization. Int J Mol Sci 21, 7875.
DOI URL |
| [82] |
Ralet MC, Crepeau MJ, Vigouroux J, Tran J, Berger A, Salle C, Granier F, Botran L, North HM (2016). Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol 171, 165-178.
DOI URL |
| [83] |
Ratke C, Pawar PMA, Balasubramanian VK, Naumann M, Duncranz ML, Derba-Maceluch M, Gorzsás A, Endo S, Ezcurra I, Mellerowicz EJ (2015). Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol J 13, 26-37.
DOI URL |
| [84] |
Ratke C, Terebieniec BK, Winestrand S, Derba-Maceluch M, Grahn T, Schiffthaler B, Ulvcrona T, Özparpucu M, Rüggeberg M, Lundqvist SO, Street NR, Jönsson LJ, Mellerowicz EJ (2018). Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol 219, 230-245.
DOI PMID |
| [85] |
Rautengarten C, Birdseye D, Pattathil S, McFarlane HE, Saez-Aguayo S, Orellana A, Persson S, Hahn MG, Scheller HV, Heazlewood JL, Ebert B (2017). The elaborate route for UDP-arabinose delivery into the Golgi of plants. Proc Natl Acad Sci USA 114, 4261-4266.
DOI PMID |
| [86] |
Rennie EA, Hansen SF, Baidoo EEK, Hadi MZ, Keasling JD, Scheller HV (2012). Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol 159, 1408-1417.
DOI URL |
| [87] |
Rennie EA, Scheller HV (2014). Xylan biosynthesis. Curr Opin Biotechnol 26, 100-107.
DOI URL |
| [88] |
Ruan N, Dang ZJ, Wang MH, Cao LY, Wang Y, Liu ST, Tang YJ, Huang YW, Zhang Q, Xu Q, Chen WF, Li FC (2022). FRAGILE CULM 18encodes a UDP-glucuronic acid decarboxylase required for xylan biosynthesis and plant growth in rice. J Exp Bot 73, 2320-2335.
DOI PMID |
| [89] |
Saez-Aguayo S, Rautengarten C, Temple H, Sanhueza D, Ejsmentewicz T, Sandoval-Ibañez O, Doñas D, Parra-Rojas JP, Ebert B, Lehner A, Mollet JC, Dupree P, Scheller HV, Heazlewood JL, Reyes FC, Orellana A (2017). UUAT1 is a Golgi-localized UDP-uronic acid transporter that modulates the polysaccharide composition of Arabidopsis seed mucilage. Plant Cell 29, 129-143.
DOI URL |
| [90] |
Schultink A, Naylor D, Dama M, Pauly M (2015). The role of the plant-specific ALTERED XYLOGLUCAN 9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol 167, 1271-1283.
DOI PMID |
| [91] |
Smith PJ, Wang HT, York WS, Peña MJ, Urbanowicz BR (2017). Designer biomass for next-generation biorefineries: leveraging recent insights into xylan structure and biosynthesis. Biotechnol Biofuels 10, 286.
DOI PMID |
| [92] | Song DL, Gui JS, Liu CC, Sun JY, Li LG (2016). Suppression of PtrDUF579-3 expression causes structural changes of the glucuronoxylan in Populus. Front Plant Sci 7, 493. |
| [93] |
Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan CH, Hao ZY, Zhu X, Avci U, Miller JS, Baldwin D, Pham C, Orlando R, Darvill A, Hahn MG, Kieliszewski MJ, Mohnen D (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270-287.
DOI URL |
| [94] |
Teleman A, Lundqvist J, Tjerneld F, Stålbrand H, Dahlman O (2000). Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr Res 329, 807-815.
DOI URL |
| [95] |
Terrett OM, Dupree P (2019). Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr Opin Biotechnol 56, 97-104.
DOI URL |
| [96] | Tucker MR, Ma C, Phan J, Neumann K, Shirley NJ, Hahn MG, Cozzolino D, Burton RA (2017). Dissecting the genetic basis for seed coat mucilage heteroxylan biosynthesis in Plantago ovata using gamma irradiation and infrared spectroscopy. Front Plant Sci 8, 326. |
| [97] |
Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014). Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80, 197-206.
DOI URL |
| [98] |
Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li HJ, O'Neill MA, Ragauskas AJ, Darvill AG, Wyman C, Gilbert HJ, York WS (2012). 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc Natl Acad Sci USA 109, 14253-14258.
DOI PMID |
| [99] |
Verbruggen MA, Spronk BA, Schols HA, Beldman G, Voragen AGJ, Thomas JR, Kamerling JP, Vliegenthart JFG (1998). Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr Res 306, 265-274.
DOI URL |
| [100] |
Wang H, Yang HL, Wen Z, Gao CX, Gao YH, Tian YB, Xu ZP, Liu XL, Persson S, Zhang BC, Zhou YH (2022). Xylan-based nanocompartments orchestrate plant vessel wall patterning. Nat Plants 8, 295-306.
DOI PMID |
| [101] |
Wang YP, Xu Y, Pei SQ, Lu MM, Kong YZ, Zhou GK, Hu RB (2020). KNAT7 regulates xylan biosynthesis in Arabidopsis seed-coat mucilage. J Exp Bot 71, 4125-4139.
DOI URL |
| [102] | Whitehead C, Garrido FJO, Reymond M, Simister R, Distelfeld A, Atienza SG, Piston F, Gomez LD, McQueen- Mason SJ (2018). A glycosyl transferase family 43 protein involved in xylan biosynthesis is associated with straw digestibility in Brachypodium distachyon. New Phytol 218, 974-985. |
| [103] | Wilkinson MD, Kosik O, Halsey K, Walpole H, Evans J, Wood AJ, Ward JL, Mitchell RAC, Lovegrove A, Shewry PR (2021). RNAi suppression of xylan synthase genes in wheat starchy endosperm. PLoS One 16, e0256350. |
| [104] |
Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57, 718-731.
DOI URL |
| [105] |
Wu X, Delbianco M, Anggara K, Michnowicz T, Pardo- Vargas A, Bharate P, Sen S, Pristl M, Rauschenbach S, Schlickum U, Abb S, Seeberger PH, Kern K (2020). Imaging single glycans. Nature 582, 375-378.
DOI |
| [106] |
Xiong GY, Cheng K, Pauly M (2013). Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6, 1373-1375.
DOI URL |
| [107] |
Yuan YX, Teng Q, Zhong RQ, Ye ZH (2013). The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 54, 1186-1199.
DOI URL |
| [108] |
Yuan YX, Teng Q, Zhong RQ, Ye ZH (2016a). Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci 243, 120-130.
DOI URL |
| [109] |
Yuan YX, Teng Q, Zhong RQ, Ye ZH (2016b). TBL3 and TBL31, two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol 57, 35-45.
DOI URL |
| [110] |
Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A (2010). A glucurono (arabino) xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154, 78-97.
DOI PMID |
| [111] |
Zeng W, Lampugnani ER, Picard KL, Song LL, Wu AM, Farion IM, Zhao J, Ford K, Doblin MS, Bacic A (2016). Asparagus IRX9, IRX10, and IRX14A are components of an active xylan backbone synthase complex that forms in the Golgi apparatus. Plant Physiol 171, 93-109.
DOI PMID |
| [112] |
Zhang BC, Gao YH, Zhang LJ, Zhou YH (2021). The plant cell wall: biosynthesis, construction, and functions. J Integr Plant Biol 63, 251-272.
DOI |
| [113] |
Zhang BC, Zhang LJ, Li F, Zhang DM, Liu XL, Wang H, Xu ZP, Chu CC, Zhou YH (2017). Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat Plants 3, 17017.
DOI PMID |
| [114] |
Zhang LJ, Gao CX, Mentink-Vigier F, Tang L, Zhang DM, Wang SG, Cao SX, Xu ZP, Liu XL, Wang T, Zhou YH, Zhang BC (2019). Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell 31, 1113-1126.
DOI URL |
| [115] | Zhao XH, Liu N, Shang N, Zeng W, Ebert B, Rautengarten C, Zeng QY, Li HL, Chen XY, Beahan C, Bacic A, Heazlewood JL, Wu AM (2018). Three UDP-xylose transporters participate in xylan biosynthesis by conveying cytosolic UDP-xylose into the Golgi lumen in Arabidopsis. J Exp Bot 69, 1125-1134. |
| [116] |
Zhong RQ, Cui DT, Dasher RL, Ye ZH (2018a). Biochemical characterization of rice xylan O-acetyltransferases. Planta 247, 1489-1498.
DOI |
| [117] |
Zhong RQ, Cui DT, Phillips DR, Sims NT, Ye ZH (2021). Functional analysis of GT61 glycosyltransferases from grass species in xylan substitutions. Planta 254, 131.
DOI PMID |
| [118] |
Zhong RQ, Cui DT, Phillips DR, Ye ZH (2018b). A novel rice xylosyltransferase catalyzes the addition of 2-O-xylosyl side chains onto the xylan backbone. Plant Cell Physiol 59, 554-565.
DOI URL |
| [119] |
Zhong RQ, Cui DT, Ye ZH (2017a). Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol 58, 2126-2138.
DOI URL |
| [120] | Zhong RQ, Cui DT, Ye ZH (2018c). A group of Populus trichocarpa DUF231 proteins exhibit differential O-acetyltransferase activities toward xylan. PLoS One 13, e0194532. |
| [121] |
Zhong RQ, Cui DT, Ye ZH (2018d). Xyloglucan O-acetyltransferases from Arabidopsis thaliana and Populus trichocarpa catalyze acetylation of fucosylated galactose residues on xyloglucan side chains. Planta 248, 1159-1171.
DOI |
| [122] |
Zhong RQ, Cui DT, Ye ZH (2019). Secondary cell wall biosynthesis. New Phytol 221, 1703-1723.
DOI PMID |
| [123] | Zhong RQ, Lee C, Ye ZH (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3, 1087-1103. |
| [124] |
Zhong RQ, Peña MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison III WH, Darvill AG, York WS, Ye ZH (2005). Arabidopsis FRAGILE FIBER 8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17, 3390-3408.
DOI URL |
| [125] | Zhong RQ, Teng Q, Haghighat M, Yuan YX, Furey ST, Dasher RL, Ye ZH (2017b). Cytosol-localized UDP-xylose synthases provide the major source of UDP-xylose for the biosynthesis of xylan and xyloglucan. Plant Cell Physiol 58, 156-174. |
| [126] |
Zhong RQ, Ye ZH (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53, 368-380.
DOI PMID |
| [127] |
Zhou GK, Zhong RQ, Himmelsbach DS, McPhail BT, Ye ZH (2007). Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiol 48, 689-699.
DOI URL |
| [128] |
Zhou GK, Zhong RQ, Richardson EA, Morrison III WH, Nairn CJ, Wood-Jones A, Ye ZH (2006). The poplar glycosyltransferase GT47C is functionally conserved with Arabidopsis Fragile Fiber 8. Plant Cell Physiol 47, 1229-1240.
DOI URL |
| [1] | 周婧, 高飞. 植物缺铁诱导型香豆素合成及其在铁吸收中的功能研究进展[J]. 植物学报, 2025, 60(3): 460-471. |
| [2] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [3] | 胡海涛, 武越, 杨玲. 植物NAD(P)+的生物合成及其生物学功能研究进展[J]. 植物学报, 2025, 60(1): 114-131. |
| [4] | 王子阳, 刘升学, 杨志蕊, 秦峰. 玉米抗旱性的遗传解析[J]. 植物学报, 2024, 59(6): 883-902. |
| [5] | 张强, 赵振宇, 李平华. 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024, 59(6): 978-998. |
| [6] | 覃思颖, 罗燕, 张禾, 胡君, 廖菊够. 花粉管细胞壁原子力显微镜观测制样方法优化[J]. 植物学报, 2024, 59(5): 783-791. |
| [7] | 范雪兰, 落艳娇, 徐超群, 郭宝林. 淫羊藿类黄酮生物合成相关基因研究进展[J]. 植物学报, 2024, 59(5): 834-846. |
| [8] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [9] | 朱璐, 袁冲, 刘义飞. 植物次生代谢产物生物合成基因簇研究进展[J]. 植物学报, 2024, 59(1): 134-143. |
| [10] | 刘潇潇, 巩迪, 高天鹏, 殷俐娜, 王仕稳. 植物类囊体主要膜脂及其生物合成[J]. 植物学报, 2024, 59(1): 144-155. |
| [11] | 张御格, 袁笑妍, 张贵芳, 李雨健, 殷金环, 林金星, 李晓娟. 点击化学反应在植物细胞标记中的应用[J]. 植物学报, 2023, 58(6): 956-965. |
| [12] | 胡海涛, 郭龙彪. 植物核黄素的生物合成及其功能研究进展[J]. 植物学报, 2023, 58(4): 638-655. |
| [13] | 冯旭飞, 雷长英, 张玉洁, 向导, 杨明凤, 张旺锋, 张亚黎. 棉花花铃期叶片氮分配对光合氮利用效率的影响[J]. 植物生态学报, 2023, 47(11): 1600-1610. |
| [14] | 熊映杰, 于果, 魏凯璐, 彭娟, 耿鸿儒, 杨冬梅, 彭国全. 天童山阔叶木本植物叶片大小与叶脉密度及单位叶脉长度细胞壁干质量的关系[J]. 植物生态学报, 2022, 46(2): 136-147. |
| [15] | 刘德帅, 姚磊, 徐伟荣, 冯美, 姚文孔. 褪黑素参与植物抗逆功能研究进展[J]. 植物学报, 2022, 57(1): 111-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||