

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (5): 594-602.DOI: 10.11983/CBB17114 cstr: 32102.14.CBB17114
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Lu Dan, Wang Li, Song Fan, Tao Juhong, Zhang Dabing, Yuan Zheng*( )
)
Received:2017-06-07
Accepted:2017-10-07
Online:2018-09-01
Published:2018-11-29
Contact:
Yuan Zheng
About author:† These authors contributed equally to this paper
Lu Dan, Wang Li, Song Fan, Tao Juhong, Zhang Dabing, Yuan Zheng. Cloning and Expression Pattern Analysis of Rice OsJMJ718 Alternative Polyadenylation Sequences During Reproductive Developmental Stage[J]. Chinese Bulletin of Botany, 2018, 53(5): 594-602.
| Primer name | Primer sequence (5′-3′) | Type |
|---|---|---|
| qRT-F-TVX1 | GAGCTTGGATAGCCCGCCTC | qRT PCR |
| qRT-R-TVX1 | TCTTTTCTTCCCGGGAGTGC | qRT PCR |
| qRT-F-TVX2 | CAATAATTGAACTCTAGGTC | qRT PCR |
| qRT-R-TVX2 | TAAGGAAATACAATCAGATC | qRT PCR |
| qRT-F-TVX3 | ACGGACGCTGGATCGGCGAG | qRT PCR |
| qRT-R-TVX3 | TAACAAGAGCAGTAGAGCAC | qRT PCR |
| qRT-F-TVX4 | ACGGACGCTGGATCGGCGAG | qRT PCR |
| qRT-R-TVX4 | AAGGACGGGGATGCGGCGT | qRT PCR |
| qRT-F-TVX5 | AACGACAACTTTAGGGTTCG | qRT PCR |
| qRT-R-TVX5 | TCGTTACAAGAAAGATGAAC | qRT PCR |
| qRT-F-TVX6 | ATCGAATTGCCACGTAAGCG | qRT PCR |
| qRT-R-TVX6 | TCATCCTCACTCTCTTCTTC | qRT PCR |
| qRT-F-TVX7 | GAACCACAGGGCCAAAGAAG | qRT PCR |
| qRT-R-TVX7 | TAATCCAATTAAAAGTGTTG | qRT PCR |
| qRT-F-TVX9 | AACTCTTCACCACGCGTATG | qRT PCR |
| qRT-R-TVX9 | TAACCGGCGATGGCTGCATC | qRT PCR |
| qRT-F-TVX11 | GAATAAGATGATAATCTATG | qRT PCR |
| qRT-R-TVX11 | ATATCTCTAACTCTACATGC | qRT PCR |
| 3'RACE-F-OsJMJ718 | GATTACGCCAAGCTTAGTGAGACCAACAAGGGAGGTGCT | 3'RACE |
Table 1 Primers used in this research
| Primer name | Primer sequence (5′-3′) | Type |
|---|---|---|
| qRT-F-TVX1 | GAGCTTGGATAGCCCGCCTC | qRT PCR |
| qRT-R-TVX1 | TCTTTTCTTCCCGGGAGTGC | qRT PCR |
| qRT-F-TVX2 | CAATAATTGAACTCTAGGTC | qRT PCR |
| qRT-R-TVX2 | TAAGGAAATACAATCAGATC | qRT PCR |
| qRT-F-TVX3 | ACGGACGCTGGATCGGCGAG | qRT PCR |
| qRT-R-TVX3 | TAACAAGAGCAGTAGAGCAC | qRT PCR |
| qRT-F-TVX4 | ACGGACGCTGGATCGGCGAG | qRT PCR |
| qRT-R-TVX4 | AAGGACGGGGATGCGGCGT | qRT PCR |
| qRT-F-TVX5 | AACGACAACTTTAGGGTTCG | qRT PCR |
| qRT-R-TVX5 | TCGTTACAAGAAAGATGAAC | qRT PCR |
| qRT-F-TVX6 | ATCGAATTGCCACGTAAGCG | qRT PCR |
| qRT-R-TVX6 | TCATCCTCACTCTCTTCTTC | qRT PCR |
| qRT-F-TVX7 | GAACCACAGGGCCAAAGAAG | qRT PCR |
| qRT-R-TVX7 | TAATCCAATTAAAAGTGTTG | qRT PCR |
| qRT-F-TVX9 | AACTCTTCACCACGCGTATG | qRT PCR |
| qRT-R-TVX9 | TAACCGGCGATGGCTGCATC | qRT PCR |
| qRT-F-TVX11 | GAATAAGATGATAATCTATG | qRT PCR |
| qRT-R-TVX11 | ATATCTCTAACTCTACATGC | qRT PCR |
| 3'RACE-F-OsJMJ718 | GATTACGCCAAGCTTAGTGAGACCAACAAGGGAGGTGCT | 3'RACE |
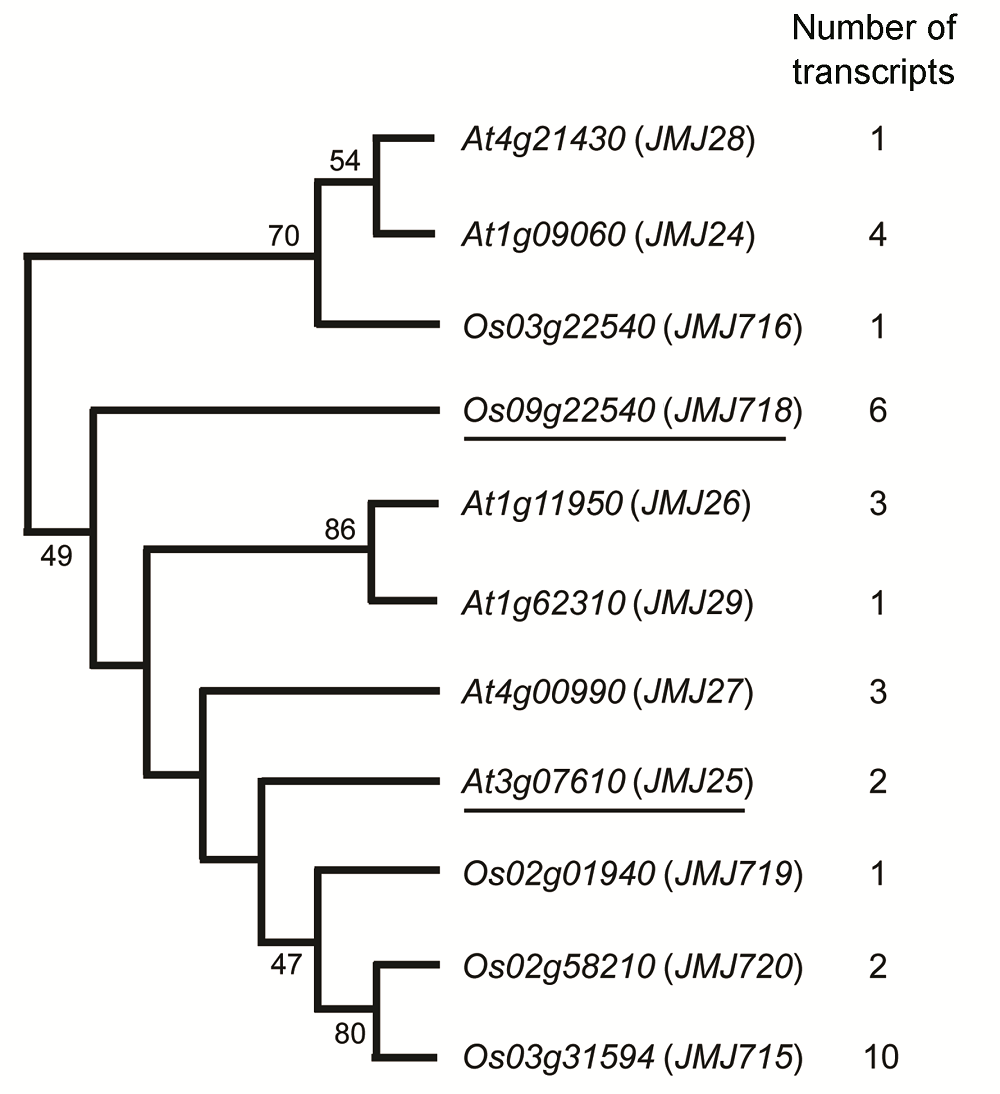
Figure 1 The number of alternative polyadenylation sequences of JHDM2 family genes in Arabidopsis thaliana and riceOs09g22540 (JMJ718) is OsJMJ718, the target sequence of this study; At3g07610 (JMJ25) is IBM1, which is the homo- logous gene of OsJMJ718 in Arabidopsis thaliana.
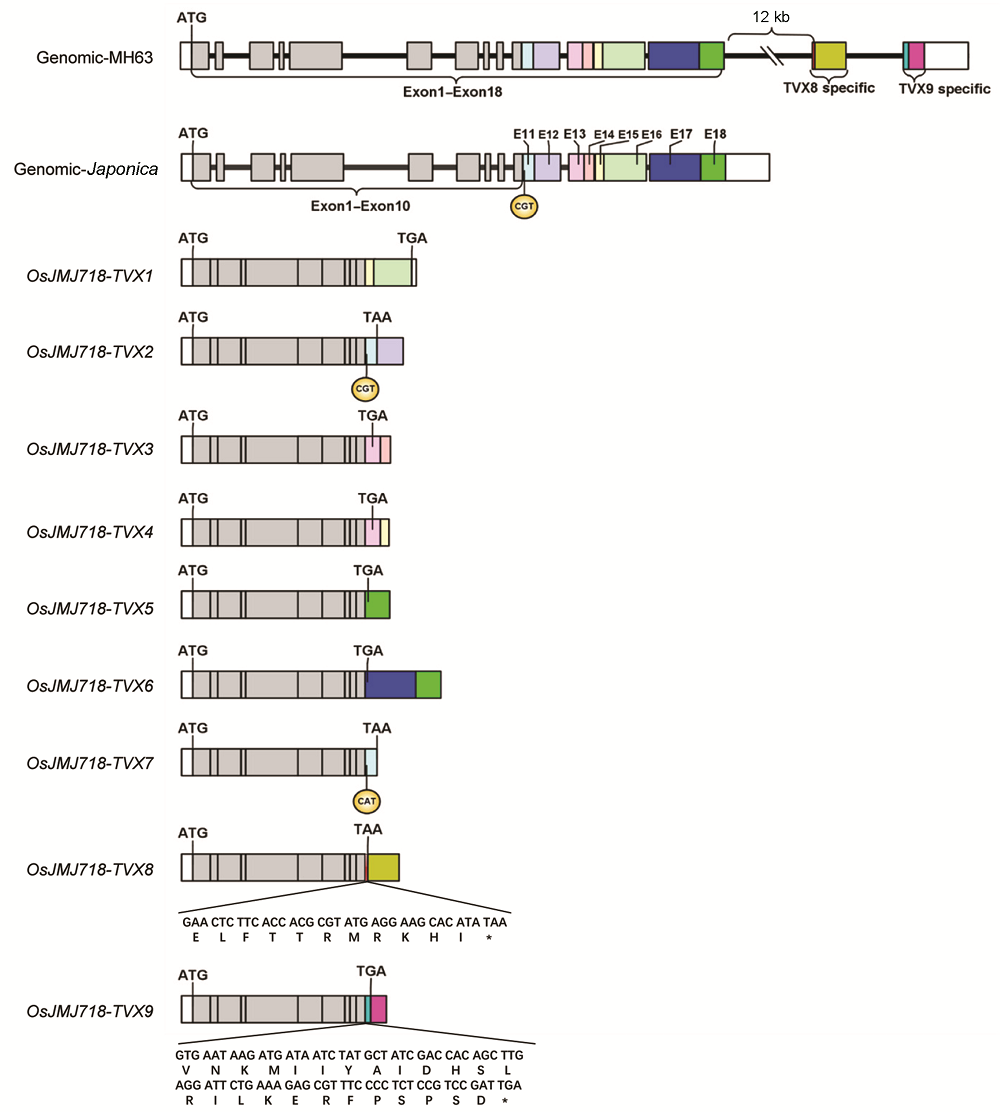
Figure 2 Sequence analysis of OsJMJ718-TVX transcriptsNine transcripts were named as TVX1-TVX9, respectively; Exon1-Exon10 represent common exon sequences for each transcript; Exon11-Exon18 were marked in different color, the 8 exons were processed in TVX1-TVX7 by alternative polyadenylation. However, 3’ terminal sequences of TVX8 and TVX9 are only exist in MH63 genomic sequences. * indicates stop codon
| 11 | Saze H, Shiraishi A, Miura A, Kakutani T (2008). Control of genic DNA methylation by a jmjC domain-containing pro- tein in Arabidopsis thaliana. Science 319, 462-465. |
| 12 | Shen YJ, Venu RC, Nobuta K, Wu XH, Notibala V, Demirci C, Meyers BC, Wang GL, Ji GL, Li QQ (2011). Transcrip- tome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing.Genome Res 21, 1478-1486. |
| 13 | Shi YS (2012). Alternative polyadenylation: new insights from global analyses.RNA 18, 2105-2117. |
| 14 | Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003). FY is an RNA 3′ end-processing factor that inter- acts with FCA to control the Arabidopsis floral transition.Cell 113, 777-787. |
| 15 | Sun Q, Zhou DX (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105, 13679-13684. |
| 16 | Wu XH, Liu M, Downie B, Liang C, Ji GL, Li QQ, Hunt AG (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation.Proc Natl Acad Sci USA 108, 12533-12538. |
| 17 | Xing DH, Li QQ (2011). Alternative polyadenylation and gene expression regulation in plants.Wiley Interdiscip Rev RNA 2, 445-458. |
| 18 | Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu YC, Yan J, Saidel GM, Fox PL (2012). Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression.Cell 149, 88-100. |
| 19 | Zhang JX, Addepalli B, Yun KY, Hunt AG, Xu RQ, Rao S, Li QQ, Falcone DL (2008). A polyadenylation factor subunit implicated in regulating oxidative signaling in Ara- bidopsis thaliana. PLoS One 3, e2410. |
| 20 | Zhang JW, Chen LL, Xing F, Kudrna DA, Yao W, Copetti D, Mu T, Li WM, Song JM, Xie WB, Lee S, Talag J, Shao L, An Y, Zhang CL, Ouyang YD, Sun S, Jiao WB, Lv F, Du BG, Luo MZ, Maldonado CE, Goicoechea JL, Xiong LZ, Wu CY, Xing YZ, Zhou DX, Yu SB, Zhao Y, Wang GW, Yu Y, Luo YJ, Zhou ZW, Hurtado BEP, Danowitz A, Wing RA, Zhang QF (2016). Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc Natl Acad Sci USA 113, E5163-E5171. |
| 1 | Elkon R, Ugalde AP, Agami R (2013). Alternative cleavage and polyadenylation: extent, regulation and function.Nat Rev Genet 14, 496-506. |
| 2 | Inagaki S, Miura-Kamio A, Nakamura Y, Lu FL, Cui X, Cao XF, Kimura H, Saze H, Kakutani T (2010). Autocatalytic differentiation of epigenetic modifications within the Ara- bidopsis genome.EMBO J 29, 3496-3506. |
| 3 | Itoh JI, Nonomura KI, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005). Rice plant development: from zygote to spikelet.Plant Cell Physiol 46, 23-47. |
| 4 | Lei MG, La HG, Lu K, Wang PC, Miki D, Ren ZZ, Duan CG, Wang XG, Tang K, Zeng L, Yang L, Zhang H, Nie WF, Liu P, Zhou JP, Liu RY, Zhong YL, Liu D, Zhu JK (2014). Arabidopsis EDM2 promotes IBM1 distal poly- adenylation and regulates genome DNA methylation pat- terns. Proc Natl Acad Sci USA 111, 527-532. |
| 5 | Licatalosi DD, Darnell RB (2010). RNA processing and its regulation: global insights into biological networks.Nat Rev Genet 11, 75-87. |
| 6 | Liu Y, Cui SJ, Wu F, Yan S, Lin XL, Du XQ, Chong K, Schilling S, Theissen G, Meng Z (2013). Functional conservation of MIKC*-type MADS box genes in Ara- bidopsis and rice pollen maturation.Plant Cell 25, 1288-1303. |
| 7 | Ma LY, Guo C, Li QQ (2014). Role of alternative poly- adenylation in epigenetic silencing and antisilencing.Proc Natl Acad Sci USA 111, 9-10. |
| 8 | Mayr C, Bartel DP (2009). Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates onco- genes in cancer cells.Cell 138, 673-684. |
| 9 | Rigal M, Kevei Z, Pélissier T, Mathieu O (2012). DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns.EMBO J 31, 2981-2993. |
| 10 | Rosonina E, Manley JL (2010). Alternative polyadenylation blooms.Dev Cell 18, 172-174. |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [3] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [4] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [5] | Qingyang Li, Cui Liu, Li He, Shan Peng, Jiayin Ma, Ziyi Hu, Hongbo Liu. Cloning and Functional Analysis of the BnaA02.CPSF6 Gene from Brassica napus [J]. Chinese Bulletin of Botany, 2025, 60(1): 62-73. |
| [6] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [7] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [8] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [9] | Lei Gu, Qi Zhang, Xia Zhang, Bingbing Yang, Fanglan Wang, Wen Liu, Faju Chen. Cloning and Functional Analysis of APETALA3/DEFICIENS Homologous Gene from Rhus chinensis [J]. Chinese Bulletin of Botany, 2024, 59(4): 533-543. |
| [10] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [11] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [12] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [13] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [14] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [15] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||