

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (2): 257-277.DOI: 10.11983/CBB23096 cstr: 32102.14.CBB23096
• SPECIAL TOPICS • Previous Articles Next Articles
Wen Chen*,†( ), Yingying Zhou,†, Ping Luo, Yongyi Cui
), Yingying Zhou,†, Ping Luo, Yongyi Cui
Received:2023-07-22
Accepted:2023-11-14
Online:2024-03-10
Published:2024-03-10
Contact:
* E-mail: About author:† These authors contributed equally to this paper
Wen Chen, Yingying Zhou, Ping Luo, Yongyi Cui. Molecular Mechanism of Petal Doubling of Flower in Angiosperm[J]. Chinese Bulletin of Botany, 2024, 59(2): 257-277.
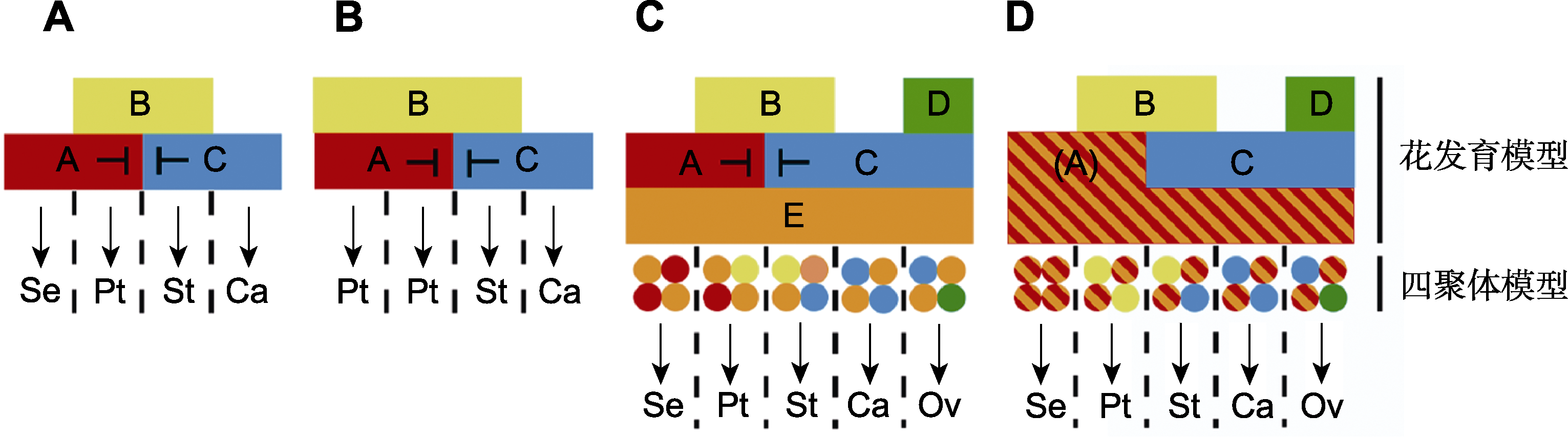
Figure 1 Flower development models of Angiosperm (refer to Soltis et al., 2007; Thomson and Wellmer, 2019) (A) Classic ABC model (represented by plants such as Arabidopsis thaliana, snapdragon and rose); (B) Modified ABC model (represented by plants of Tulipa, Lilium, Ranunculus and Aquilegia); (C) ABCDE model (represented by model plants such as Arabidopsis thaliana and snapdragon); (D) (A)BCD model (represented by plants such as gerbera and rice, the defined (A) combines A and E function genes). Arrows represent the regulation of functional genes on specific floral organs; T-shaped lines represent repression. Se: Sepals; Pt: Petals; St: Stamens; Ca: Carpels; Ov: Ovules
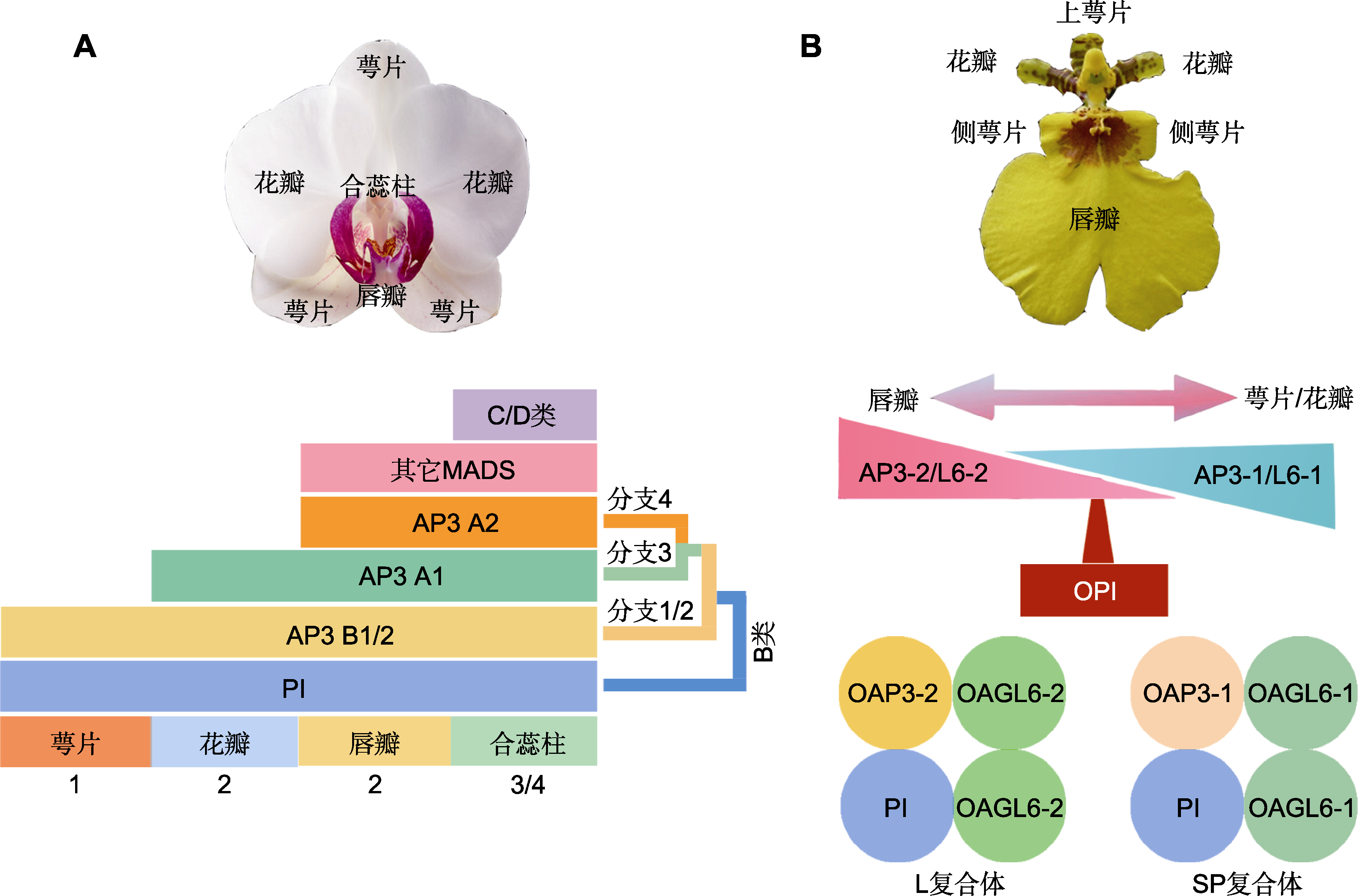
Figure 2 HOT model and perianth code for flower development of orchids (refer to Hsu et al., 2015; Zhang et al., 2022) (A) HOT model for flower development of orchids (PI and four clades of AP3 of B-class genes specify the orchid perianth formation at the late inflorescence and floral bud stages. At the late inflorescence stage, PI and AP3Bs determine the formation of sepals. PI and both AP3A1 and AP3Bs control the lateral petal formation; at the floral bud stage, PI, AP3As and other MADS-box genes control the lip morphogenesis); (B) Perianth code of orchids (in the sepal, petal and lip, PI interacts with different AP3-like and AGL-like proteins to form L complex (OAP3-2/OAGL6-2/OAGR6-2/OPI) or SP complex (OAP3-1/OAGL6-1/OGL6-1/OPI). The presence of L complex results in lip formation, and the presence of SP complex results in sepal/petal formation; the coexistence or co-absence of the SP and L complexes produces sepal/petal and lip intermediate structures). Numbers 1, 2, 3/4 represent flower whorls.
| 类型 | 名称 | 功能及表型 | 重瓣起源类型 | 参考文献 |
|---|---|---|---|---|
| A类功能基因 | AP2 | 月季RcAP2表达水平受低温诱导而被高温抑制, 拟南芥异源过表达RcAP2可诱导雄蕊瓣化 | 雌雄蕊起源 | Han et al., |
| B类功能基因(MADS-box) | PI | 拟南芥中异源过表达桃PpPI可诱导雄蕊瓣化; 拟南芥中异源过表达枇杷EjPI可导致萼片瓣化 | 雌雄蕊起源或苞片起源 | Xia et al., |
| AP3 | 夏堇中共表达TfDEF (AP3)和TfGLO (PI)可导致萼片瓣化; 共抑制两者可导致花瓣萼片化 | 苞片起源 | Sasaki et al., | |
| C类功能基因(MADS-box) | AG | 矮牵牛、三花龙胆和月季中沉默AG同源基因导致雄蕊瓣化; 低温诱导的月季重瓣化可能与RhAG表达区域被低温抑制有关 | 雌雄蕊起源 | Noor et al., |
| E类功能基因(MADS-box) | SEP3 | 莲NnSEP3过表达导致花萼和花瓣数量减少, 类似心皮的器官增加 | 雌雄蕊起源 | 林钟员, |
| SEP4 | 万寿菊TeSEP4过表达导致花瓣和雄蕊数量减少, 该蛋白可与多种MADS-box蛋白互作 | 雌雄蕊起源 | Zhang et al., | |
| 干细胞决定基因 | WUS | WUS与CLV可形成负反馈回路调控花分生组织活性的终止。WUS表达受AG和SEP抑制。WUS功能缺失导致干细胞和花器官数量减少 | 其它 | Ito et al., |
| 边界基因 | PTL | PTL可通过维持生长素平衡确保花瓣正常发育。拟南芥PTL缺失可导致花瓣减少。PTL抑制C类功能基因在第1、2轮花器官的表达 | 其它 | Lampugnani et al., |
| HAN | HAN可通过维持边界细胞中细胞分裂素的平衡而调控花器官发育。其突变导致拟南芥萼片融合及花瓣和雄蕊数量减少 | 其它 | Zhao et al., | |
| CUC | CUCs在花器官边界表达并抑制细胞增殖。CUC1和CUC2基因受miRNA164a-c负调控, CUCs双重突变导致萼片融合和花瓣数量减少 | 其它 | Aida et al., | |
| SUP | SUP主要控制第3轮与第4轮花器官之间的边界, 可能影响生长素合成。sup/ag-1双突变体在心皮位置较ag-1具有更多轮次花瓣的重瓣性状 | 其它 | Prunet et al., | |
| RBE | RBE可通过抑制AG表达而控制第2轮与第3轮花器官边界。其突变导致花瓣被雄蕊或雄蕊类似器官替代 | 其它 | Krizek et al., | |
| 对称性基因 | CYC | 向日葵和非洲菊CYC同源基因的异位表达导致管状花向舌状花转变, 并通过调控细胞增殖影响舌状花花瓣的形状和大小 | 花序起源 | Fambrini et al., |
| 其它 | MYB | 月季中沉默RhMYB123导致雄蕊瓣化, RhAG、RhAGL15和RhSHP1及生长素信号转导途径中多个基因的表达量下降 | 雌雄蕊起源 | Li et al., |
| PAN | 拟南芥pan突变体前三轮花器官的数量均增加。其可能通过控制花器官原基起始时间间隔影响花器官的数量和位置 | 其它 | Running and Meyerowitz, | |
| ULT | 拟南芥中ULT1突变导致4轮花器官数量均增多。其可能通过调节WUS-AG反馈环参与干细胞的终止 | 其它 | Fletcher, |
Table 1 Key transcription factor genes and their functions involved in petal doubling of flower
| 类型 | 名称 | 功能及表型 | 重瓣起源类型 | 参考文献 |
|---|---|---|---|---|
| A类功能基因 | AP2 | 月季RcAP2表达水平受低温诱导而被高温抑制, 拟南芥异源过表达RcAP2可诱导雄蕊瓣化 | 雌雄蕊起源 | Han et al., |
| B类功能基因(MADS-box) | PI | 拟南芥中异源过表达桃PpPI可诱导雄蕊瓣化; 拟南芥中异源过表达枇杷EjPI可导致萼片瓣化 | 雌雄蕊起源或苞片起源 | Xia et al., |
| AP3 | 夏堇中共表达TfDEF (AP3)和TfGLO (PI)可导致萼片瓣化; 共抑制两者可导致花瓣萼片化 | 苞片起源 | Sasaki et al., | |
| C类功能基因(MADS-box) | AG | 矮牵牛、三花龙胆和月季中沉默AG同源基因导致雄蕊瓣化; 低温诱导的月季重瓣化可能与RhAG表达区域被低温抑制有关 | 雌雄蕊起源 | Noor et al., |
| E类功能基因(MADS-box) | SEP3 | 莲NnSEP3过表达导致花萼和花瓣数量减少, 类似心皮的器官增加 | 雌雄蕊起源 | 林钟员, |
| SEP4 | 万寿菊TeSEP4过表达导致花瓣和雄蕊数量减少, 该蛋白可与多种MADS-box蛋白互作 | 雌雄蕊起源 | Zhang et al., | |
| 干细胞决定基因 | WUS | WUS与CLV可形成负反馈回路调控花分生组织活性的终止。WUS表达受AG和SEP抑制。WUS功能缺失导致干细胞和花器官数量减少 | 其它 | Ito et al., |
| 边界基因 | PTL | PTL可通过维持生长素平衡确保花瓣正常发育。拟南芥PTL缺失可导致花瓣减少。PTL抑制C类功能基因在第1、2轮花器官的表达 | 其它 | Lampugnani et al., |
| HAN | HAN可通过维持边界细胞中细胞分裂素的平衡而调控花器官发育。其突变导致拟南芥萼片融合及花瓣和雄蕊数量减少 | 其它 | Zhao et al., | |
| CUC | CUCs在花器官边界表达并抑制细胞增殖。CUC1和CUC2基因受miRNA164a-c负调控, CUCs双重突变导致萼片融合和花瓣数量减少 | 其它 | Aida et al., | |
| SUP | SUP主要控制第3轮与第4轮花器官之间的边界, 可能影响生长素合成。sup/ag-1双突变体在心皮位置较ag-1具有更多轮次花瓣的重瓣性状 | 其它 | Prunet et al., | |
| RBE | RBE可通过抑制AG表达而控制第2轮与第3轮花器官边界。其突变导致花瓣被雄蕊或雄蕊类似器官替代 | 其它 | Krizek et al., | |
| 对称性基因 | CYC | 向日葵和非洲菊CYC同源基因的异位表达导致管状花向舌状花转变, 并通过调控细胞增殖影响舌状花花瓣的形状和大小 | 花序起源 | Fambrini et al., |
| 其它 | MYB | 月季中沉默RhMYB123导致雄蕊瓣化, RhAG、RhAGL15和RhSHP1及生长素信号转导途径中多个基因的表达量下降 | 雌雄蕊起源 | Li et al., |
| PAN | 拟南芥pan突变体前三轮花器官的数量均增加。其可能通过控制花器官原基起始时间间隔影响花器官的数量和位置 | 其它 | Running and Meyerowitz, | |
| ULT | 拟南芥中ULT1突变导致4轮花器官数量均增多。其可能通过调节WUS-AG反馈环参与干细胞的终止 | 其它 | Fletcher, |
| 类型 | 作用机制 | 相关基因及其在重瓣化进程中的功能 | 参考文献 |
|---|---|---|---|
| DNA甲基化 | 常发生在CG、CHG和CHH (H包括A、T或C)的胞嘧啶C上, 影响染色质结构、DNA构象及基因表达 | 拟南芥甲基转移酶MET1缺失导致基因组DNA总体甲基化水平下降, 引起花器官同源异型转化及数量变化 | Finnegan et al., |
| 低温可通过诱导月季RhAG启动子中CHH位点甲基化水平升高而抑制RhAG表达, 导致月季雄蕊瓣化 | Ma et al., | ||
| 组蛋白甲基化 | 在转录调节和异染色质形成中发挥重要作用。通常将H3K9和H3K27甲基化作为抑制标记, 而H3K4以及H3K36甲基化则作为激活标记 | 水稻去甲基酶JMJ706功能丧失导致H3K9甲基化程度增强, 影响花的形态和器官数量 | Sun and Zhou, |
| 拟南芥AG蛋白能够通过调控WUS或KNU基因位点的H3K27甲基化修饰状态抑制WUS表达, 进而调控花分生组织的发育 | Liu et al., | ||
| TrxG蛋白家族可调控H3K4甲基化, 其家族成员的突变导致拟南芥雄蕊和花瓣间的同源异型转变 | Grini et al., | ||
| 组蛋白乙酰化 | 受组蛋白乙酰转移酶和脱乙酰酶调控。以赖氨酸乙酰化较常见, 其通常具有激活靶基因的作用 | AP2能够招募组蛋白脱乙酰化酶复合物TPL-HDA19而抑制AG的表达 | Krogan et al., |
| 月季生长素响应因子RhARF18能招募组蛋白脱乙酰酶RhHDA6到RhAG启动子上, 降低H3K9/K14乙酰化水平进而抑制其转录, 引起雄蕊瓣化 | Chen et al., | ||
| 染色质重塑 | 包括核小体位置改变、核小体去组装及组蛋白修饰等, 需要ATP依赖的染色质重塑复合物参与 | 拟南芥染色质重塑SWI/SNF复合物成员SYD和BRM基因突变均导致第2、3轮花器官的同源异型转化。SYD和BRM均能诱导AP3和AG基因表达从而启动花器官身份决定 | Hurtado et al., |
| miRNA调控 | 通过切割靶基因mRNA、抑制其翻译以及介导DNA甲基化等方式调控基因的表达 | 在桃、香石竹、矮牵牛、月季和玫瑰等植物中发现miRNA172能够抑制AP2类基因的表达进而影响花器官形态建成。该调控机制在真双子叶植物中保守 | Gattolin et al., |
| 拟南芥miR164能够通过调控边界基因CUCs的表达影响花器官数量。草莓中miRNA164-CUC也参与花型调控 | Laufs et al., | ||
| 金鱼草和矮牵牛miR169通过对NF-YA的转录后抑制降低C类基因的活性。miR169突变导致C类基因异位表达, 进而产生雄蕊化的花瓣 | Cartolano et al., | ||
| 山茶miRNA156可负调控CjSPL1/2表达, 使其仅在雄蕊花丝而不在花药中表达, 进而引起雄蕊瓣化 | Li et al., |
Table 2 Epigenetic regulations and mechanisms involved in petal doubling of flower
| 类型 | 作用机制 | 相关基因及其在重瓣化进程中的功能 | 参考文献 |
|---|---|---|---|
| DNA甲基化 | 常发生在CG、CHG和CHH (H包括A、T或C)的胞嘧啶C上, 影响染色质结构、DNA构象及基因表达 | 拟南芥甲基转移酶MET1缺失导致基因组DNA总体甲基化水平下降, 引起花器官同源异型转化及数量变化 | Finnegan et al., |
| 低温可通过诱导月季RhAG启动子中CHH位点甲基化水平升高而抑制RhAG表达, 导致月季雄蕊瓣化 | Ma et al., | ||
| 组蛋白甲基化 | 在转录调节和异染色质形成中发挥重要作用。通常将H3K9和H3K27甲基化作为抑制标记, 而H3K4以及H3K36甲基化则作为激活标记 | 水稻去甲基酶JMJ706功能丧失导致H3K9甲基化程度增强, 影响花的形态和器官数量 | Sun and Zhou, |
| 拟南芥AG蛋白能够通过调控WUS或KNU基因位点的H3K27甲基化修饰状态抑制WUS表达, 进而调控花分生组织的发育 | Liu et al., | ||
| TrxG蛋白家族可调控H3K4甲基化, 其家族成员的突变导致拟南芥雄蕊和花瓣间的同源异型转变 | Grini et al., | ||
| 组蛋白乙酰化 | 受组蛋白乙酰转移酶和脱乙酰酶调控。以赖氨酸乙酰化较常见, 其通常具有激活靶基因的作用 | AP2能够招募组蛋白脱乙酰化酶复合物TPL-HDA19而抑制AG的表达 | Krogan et al., |
| 月季生长素响应因子RhARF18能招募组蛋白脱乙酰酶RhHDA6到RhAG启动子上, 降低H3K9/K14乙酰化水平进而抑制其转录, 引起雄蕊瓣化 | Chen et al., | ||
| 染色质重塑 | 包括核小体位置改变、核小体去组装及组蛋白修饰等, 需要ATP依赖的染色质重塑复合物参与 | 拟南芥染色质重塑SWI/SNF复合物成员SYD和BRM基因突变均导致第2、3轮花器官的同源异型转化。SYD和BRM均能诱导AP3和AG基因表达从而启动花器官身份决定 | Hurtado et al., |
| miRNA调控 | 通过切割靶基因mRNA、抑制其翻译以及介导DNA甲基化等方式调控基因的表达 | 在桃、香石竹、矮牵牛、月季和玫瑰等植物中发现miRNA172能够抑制AP2类基因的表达进而影响花器官形态建成。该调控机制在真双子叶植物中保守 | Gattolin et al., |
| 拟南芥miR164能够通过调控边界基因CUCs的表达影响花器官数量。草莓中miRNA164-CUC也参与花型调控 | Laufs et al., | ||
| 金鱼草和矮牵牛miR169通过对NF-YA的转录后抑制降低C类基因的活性。miR169突变导致C类基因异位表达, 进而产生雄蕊化的花瓣 | Cartolano et al., | ||
| 山茶miRNA156可负调控CjSPL1/2表达, 使其仅在雄蕊花丝而不在花药中表达, 进而引起雄蕊瓣化 | Li et al., |
| [1] | Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841-857. |
| [2] | Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15, 303-315. |
| [3] |
Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su YH, Wagner D (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19, 403-416.
PMID |
| [4] | Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2, 615-619. |
| [5] |
Bollier N, Sicard A, Leblond J, Latrasse D, Gonzalez N, Gévaudant F, Benhamed M, Raynaud C, Lenhard M, Chevalier C, Hernould M, Delmas F (2018). At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83-100.
DOI URL |
| [6] |
Bowman JL (1997). Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J Biosci 22, 515-527.
DOI URL |
| [7] | Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599-615. |
| [8] | Bowman JL, Smyth DR, Meyerowitz EM (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1-20. |
| [9] | Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617-619. |
| [10] |
Breuil-Broyer S, Morel P, De Almeida-Engler J, Coustham V, Negrutiu I, Trehin C (2004). High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J 38, 182-192.
PMID |
| [11] | Brewer PB, Howles PA, Dorian K, Griffith ME, Ishida T, Kaplan-Levy RN, Kilinc A, Smyth DR (2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131, 4035-4045. |
| [12] |
Broholm SK, Pöllänen E, Ruokolainen S, Tähtiharju S, Kotilainen M, Albert VA, Elomaa P, Teeri TH (2010). Functional characterization of B class MADS-box transcription factors in Gerbera hybrida. J Exp Bot 61, 75-85.
DOI PMID |
| [13] | Cai YM, Wang L, Ogutu CO, Yang QR, Luo BW, Liao L, Zheng BB, Zhang RX, Han YP (2021). The MADS-box gene PpPI is a key regulator of the double-flower trait in peach. Physiol Plantarum 173, 2119-2129. |
| [14] |
Carles CC, Lertpiriyapong K, Reville K, Fletcher JC (2004). The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167, 1893-1903.
PMID |
| [15] |
Cartolano M, Castillo R, Efremova N, Kuckenberg M, Zethof J, Gerats T, Schwarz-Sommer Z, Vandenbussche M (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet 39, 901-905.
PMID |
| [16] |
Causier B, Schwarz-Sommer Z, Davies B (2010). Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21, 73-79.
DOI PMID |
| [17] |
Chapman MA, Leebens-Mack JH, Burke JM (2008). Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol Biol Evol 25, 1260-1273.
DOI PMID |
| [18] |
Chen J, Shen CZ, Guo YP, Rao GY (2018). Patterning the Asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like genes. Front Plant Sci 9, 551.
DOI PMID |
| [19] |
Chen JW, Li Y, Li YH, Li YQ, Wang Y, Jiang CY, Choisy P, Xu T, Cai YM, Pei D, Jiang CZ, Gan SS, Gao JP, Ma N (2021). AUXIN RESPONSE FACTOR 18-HISTONE DEACETYLASE 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol 186, 1074-1087.
DOI PMID |
| [20] | Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33-42. |
| [21] |
Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13, 334-344.
DOI URL |
| [22] | Coen ES, Meyerowitz EM (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31-37. |
| [23] | Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111, 14619-14624. |
| [24] |
Davie JR, Chadee DN (1998). Regulation and regulatory parameters of histone modifications. J Cell Biochem 72, 203-213.
DOI URL |
| [25] |
Deal RB, Henikoff S (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell 18, 1030-1040.
DOI PMID |
| [26] | Ding L, Yan SS, Jiang L, Zhao WS, Ning K, Zhao JY, Liu XF, Zhang J, Wang Q, Zhang XL (2015). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ONPETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet 11, e1005479. |
| [27] |
Dubois A, Raymond O, Maene M, Baudino S, Langlade NB, Boltz V, Vergne P, Bendahmane M (2010). Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One 5, e9288.
DOI URL |
| [28] | Dyson MH, Rose S, Mahadevan LC (2001). Acetyllysine- binding and function of bromodomain-containing proteins in chromatin. Front Biosci 6, 853-865. |
| [29] |
Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, Mccune RA, Gehrke C (1982). Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res 10, 2709-2721.
DOI PMID |
| [30] |
Fambrini M, Salvini M, Basile A, Pugliesi C (2014). Transposon-dependent induction of Vincent van Gogh’s sunflowers: exceptions revealed. Genesis 52, 315-327.
DOI PMID |
| [31] | Fambrini M, Salvini M, Pugliesi C (2011). A transposon- mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus annuus. Genetica 139, 1521-1529. |
| [32] |
Finnegan EJ, Peacock WJ, Dennis ES (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93, 8449-8454.
DOI PMID |
| [33] | Fletcher JC (2001). The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development 128, 1323-1333. |
| [34] |
Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911-1914.
DOI PMID |
| [35] | Gan ES, Huang JB, Ito T (2013). Functional roles of histone modification, chromatin remodeling and microRNAs in Arabidopsis flower development. Int Rev Cell Mol Biol 305, 115-161. |
| [36] | Garcês HMP, Spencer VMR, Kim M (2016). Control of floret symmetry by RAY3, SvDIV1B, and SvRAD in the capitulum of Senecio vulgaris. Plant Physiol 171, 2055-2068. |
| [37] |
Gattolin S, Cirilli M, Chessa S, Stella A, Bassi D, Rossini L (2020). Mutations in orthologous PETALOSA TOE-type genes cause a dominant double-flower phenotype in phylogenetically distant eudicots. J Exp Bot 71, 2585-2595.
DOI PMID |
| [38] |
Gattolin S, Cirilli M, Pacheco I, Ciacciulli A, Da Silva Linge C, Mauroux JB, Lambert P, Cammarata E, Bassi D, Pascal T, Rossini L (2018). Deletion of the miR172 target site in a TOE-type gene is a strong candidate variant for dominant double-flower trait in Rosaceae. Plant J 96, 358-371.
DOI URL |
| [39] | Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jørstad TS, Wilson ZA, Aalen RB (2009). The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One 4, e7817. |
| [40] |
Gul H, Tong ZG, Han XL, Nawaz I, Wahocho SA, Khan S, Zhang CX, Tian Y, Cong PH, Zhang LY (2019). Comparative transcriptome analysis between ornamental apple species provides insights into mechanism of double flowering. Agronomy 9, 112.
DOI URL |
| [41] |
Han Y, Tang AY, Wan HH, Zhang TX, Cheng TR, Wang J, Yang WR, Pan HT, Zhang QX (2018). An APETALA2 homolog, RcAP2, regulates the number of rose petals derived from stamens and response to temperature fluctuations. Front Plant Sci 9, 481.
DOI PMID |
| [42] |
Henschel K, Kofuji R, Hasebe M, Saedler H, Münster T, Theißen G (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol Biol Evol 19, 801-814.
PMID |
| [43] | Hibrand Saint-Oyant L, Ruttink T, Hamama L, Kirov I, Lakhwani D, Zhou NN, Bourke PM, Daccord N, Leus L, Schulz D, Van de Geest H, Hesselink T, Van Laere K, Debray K, Balzergue S, Thouroude T, Chastellier A, Jeauffre J, Voisine L, Gaillard S, Borm TJA, Arens P, Voorrips RE, Maliepaard C, Neu E, Linde M, Le Paslier MC, Bérard A, Bounon R, Clotault J, Choisne N, Quesneville H, Kawamura K, Aubourg S, Sakr S, Smulders MJM, Schijlen E, Bucher E, Debener T, De Riek J, Foucher F (2018). A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat Plants 4, 473-484. |
| [44] |
Hileman LC (2014). Bilateral flower symmetry—how, when and why? Curr Opin Plant Biol 17, 146-152.
DOI PMID |
| [45] |
Hileman LC, Irish VF (2009). More is better: the uses of developmental genetic data to reconstruct perianth evolution. Am J Bot 96, 83-95.
DOI PMID |
| [46] |
Hsu HF, Hsu WH, Lee YI, Mao WT, Yang JY, Li JY, Yang CH (2015). Model for perianth formation in orchids. Nat Plants 1, 15046.
DOI |
| [47] | Hu L, Zheng TC, Cai M, Pan HT, Wang J, Zhang QX (2019). Transcriptome analysis during floral organ development provides insights into stamen petaloidy in Lagerstroemia speciosa. Plant Physiol Biochem 142, 510-518. |
| [48] |
Huang TB, Irish VF (2015). Temporal control of plant organ growth by TCP transcription factors. Curr Biol 25, 1765-1770.
DOI PMID |
| [49] |
Huang TB, Irish VF (2016). Gene networks controlling petal organogenesis. J Exp Bot 67, 61-68.
DOI PMID |
| [50] |
Huang TB, López-Giráldez F, Townsend JP, Irish VF (2012). RBE controls microRNA164 expression to effect floral organogenesis. Development 139, 2161-2169.
DOI PMID |
| [51] |
Huang ZG, Shi T, Zheng BL, Yumul RE, Liu XG, You CJ, Gao ZH, Xiao LT, Chen XM (2017). APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol 215, 1197-1209.
DOI PMID |
| [52] |
Hurtado L, Farrona S, Reyes JC (2006). The putative SWI/ SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol 62, 291-304.
PMID |
| [53] |
Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM (2007). The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19, 3516-3529.
DOI PMID |
| [54] | Jacobsen SE, Meyerowitz EM (1997). Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277, 1100-1103. |
| [55] |
Jarillo JA, Piñeiro M, Cubas P, Martínez-Zapater JM (2009). Chromatin remodeling in plant development. Int J Dev Biol 53, 1581-1596.
DOI PMID |
| [56] | Jiang S, Yi XW, Xu TL, Yang Y, Yu C, Luo L, Cheng TR, Wang J, Zhang QX, Pan HT (2021). Genetic analysis of petal number in Rosa. Plant Sci J 39, 142-151. (in Chinese) |
| 姜珊, 易星湾, 徐庭亮, 杨艺, 于超, 罗乐, 程堂仁, 王佳, 张启翔, 潘会堂 (2021). 月季花瓣数量遗传分析. 植物科学学报 39, 142-151. | |
| [57] | Jing DL, Guo QG, Chen WW, Xia Y, Wu D, Dang JB, He Q, Liang GL (2018). Model evolution and molecular mechanism of angiosperm flower development. Plant Physiol J 54, 355-362. (in Chinese) |
| 景丹龙, 郭启高, 陈薇薇, 夏燕, 吴頔, 党江波, 何桥, 梁国鲁 (2018). 被子植物花器官发育的模型演变和分子调控. 植物生理学报 54, 355-362. | |
| [58] |
Jones-Rhoades MW, Bartel DP (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14, 787-799.
DOI PMID |
| [59] |
Juntheikki-Palovaara I, Tähtiharju S, Lan TY, Broholm SK, Rijpkema AS, Ruonala R, Kale L, Albert VA, Teeri TH, Elomaa P (2014). Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J 79, 783-796.
DOI URL |
| [60] | Kagale S, Rozwadowski K (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141-146. |
| [61] |
Kanno A, Saeki H, Kameya T, Saedler H, Theissen G (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52, 831-841.
DOI URL |
| [62] |
Kaufmann K, Melzer R, Theißen G (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183-198.
DOI PMID |
| [63] |
Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL (2010). Orchestration of floral initiation by APETALA1. Science 328, 85-89.
DOI PMID |
| [64] | Kermani MJ, Sarasan V, Roberts AV, Yokoya K, Wentworth J, Sieber VK (2003). Oryzalin-induced chromosome doubling in Rosa and its effect on plant morphology and pollen viability. Theor Appl Genet 107, 1195-1200. |
| [65] | Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322, 1116-1119. |
| [66] | Kramer EM, Di Stilio VS, Schluter PM (2003). Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int J Plant Sci 164, 1-11. |
| [67] | Krizek BA, Fletcher JC (2005). Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6, 688-698. |
| [68] |
Krizek BA, Lewis MW, Fletcher JC (2006). RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J 45, 369-383.
PMID |
| [69] | Krogan NT, Hogan K, Long JA (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180-4190. |
| [70] |
Kwon CS, Chen CB, Wagner D (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19, 992-1003.
DOI URL |
| [71] |
Lampugnani ER, Kilinc A, Smyth DR (2012). PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J 71, 724-735.
DOI URL |
| [72] | Lampugnani ER, Kilinc A, Smyth DR (2013). Auxin controls petal initiation in Arabidopsis. Development 140, 185-194. |
| [73] | Laufs P, Peaucelle A, Morin H, Traas J (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311-4322. |
| [74] | Laux T, Mayer KF, Berger J, Jürgens G (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87-96. |
| [75] | Lenhard M, Bohnert A, Jürgens G, Laux T (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805-814. |
| [76] |
Li K, Li YQ, Wang Y, Li YH, He JN, Li YJ, Du LS, Gao YR, Ma N, Gao JP, Zhou XF (2022). Disruption of transcription factor RhMYB123 causes the transformation of stamen to malformed petal in rose (Rosa hybrida). Plant Cell Rep 41, 2293-2303.
DOI PMID |
| [77] | Li W, Xu QJ (2014). Epigenetic research progress on flowering time and flower organ development in angiosperms. Acta Hortic Sin 41, 1245-1256. (in Chinese) |
| 李巍, 徐启江 (2014). 被子植物开花时间和花器官发育的表观遗传调控研究进展. 园艺学报 41, 1245-1256. | |
| [78] |
Li X, Qin GJ, Chen ZL, Gu HY, Qu LJ (2008). A gain-of- function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol Biol 66, 315-327.
DOI URL |
| [79] |
Li XL, Li JY, Fan ZQ, Liu ZC, Tanaka T, Yin HF (2017). Global gene expression defines faded whorl specification of double flower domestication in Camellia. Sci Rep 7, 3197.
DOI PMID |
| [80] | Lin ZY (2019). Studies on the regulatory mechanism of floral organ petaloid in lotus (Nelumbo nucifera). Doctoral dissertation. Beijing: University of Chinese Academy of Sciences (Wuhan Botanical Garden, Chinese Academy of Sciences). pp. 13-86. (in Chinese) |
| 林钟员 (2019). 莲的花器官瓣化分子调控机制的研究. 博士论文. 北京: 中国科学院大学(中国科学院武汉植物园). pp. 13-86. | |
| [81] |
Liu CY, Lu FL, Cui X, Cao XF (2010). Histone methylation in higher plants. Annu Rev Plant Biol 61, 395-420.
DOI PMID |
| [82] | Liu H, Sun M, Du DL, Pan HT, Cheng TR, Wang J, Zhang QX, Gao YK (2016). Whole-transcriptome analysis of differentially expressed genes in the ray florets and disc florets of Chrysanthemum morifolium. BMC Penomics 17, 398. |
| [83] | Liu JY, Fu XD, Dong YW, Lu J, Ren M, Zhou NN, Wang CQ (2018). MIKCC-type MADS-box genes in Rosa chinensis: the remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic Res 5, 25. |
| [84] |
Liu XG, Kim YJ, Müller R, Yumul RE, Liu CY, Pan YY, Cao XF, Goodrich J, Chen XM (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb group proteins. Plant Cell 23, 3654-3670.
DOI URL |
| [85] | Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793-803. |
| [86] | Luo Y, Guo ZH, Li L (2013). Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol 380, 133-144. |
| [87] |
Ma N, Chen W, Fan TG, Tian YR, Zhang S, Zeng DX, Li YH (2015). Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol 15, 237.
DOI PMID |
| [88] |
Mandaokar A, Browse J (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149, 851-862.
DOI PMID |
| [89] |
Martínez-Gómez J, Galimba KD, Coté EY, Sullivan AM, Di Stilio VS (2021). Spontaneous homeotic mutants and genetic control of floral organ identity in a ranunculid. Evol Dev 23, 197-214.
DOI PMID |
| [90] |
Mizzotti C, Fambrini M, Caporali E, Masiero S, Pugliesi C (2015). A CYCLOIDEA-like gene mutation in sunflower determines an unusual floret type able to produce filled achenes at the periphery of the pseudanthium. Botany 93, 171-181.
DOI URL |
| [91] |
Muiño JM, de Bruijn S, Pajoro A, Geuten K, Vingron M, Angenent GC, Kaufmann K (2016). Evolution of DNA- binding sites of a floral master regulatory transcription factor. Mol Biol Evol 33, 185-200.
DOI PMID |
| [92] |
Nakatsuka T, Saito M, Yamada E, Fujita K, Yamagishi N, Yoshikawa N, Nishihara M (2015). Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol 15, 182.
DOI PMID |
| [93] |
Ng HH, Feng Q, Wang HB, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K (2002). Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16, 1518-1527.
DOI URL |
| [94] |
Ng K, Yu H, Ito T (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol 7, e1000251.
DOI URL |
| [95] |
Ning GG, Shi XP, Hu HR, Yan Y, Bao MZ (2009). Development of a range of polyploid lines in Petunia hybrida and the relationship of ploidy with the single-/double-flower trait. HortScience 44, 250-255.
DOI URL |
| [96] | Noor SH, Ushijima K, Murata A, Yoshida K, Tanabe M, Tanigawa T, Kubo Y, Nakano R (2014). Double flower formation induced by silencing of C-class MADS-box genes and its variation among petunia cultivars. Sci Hortic 178, 1-7. |
| [97] |
Ó’Maoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwaśniewska K, Das P, Lohan AJ, Loftus B, Graciet E, Wellmer F (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25, 2482-2503.
DOI URL |
| [98] | Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, Ó’Maoiléidigh DS, Wellmer F, Krajewski P, Riechmann JL, Angenent GC, Kaufmann K (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 15, 1-19. |
| [99] |
Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, Hu JM, Chen HH (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol 52, 1515-1531.
DOI URL |
| [100] | Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200-203. |
| [101] |
Peng LP, Li Y, Tan WQ, Wu SW, Hao Q, Tong NN, Wang ZY, Liu Z, Shu QY (2023). Combined genome-wide association studies and expression quantitative trait locus analysis uncovers a genetic regulatory network of floral organ number in a tree peony (Paeonia suffruticosa Andrews) breeding population. Hortic Res 10, uhad110.
DOI URL |
| [102] |
Pien S, Grossniklaus U (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta 1769, 375-382.
PMID |
| [103] |
Prunet N, Morel P, Thierry AM, Eshed Y, Bowman JL, Negrutiu I, Trehin C (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20, 901-919.
DOI PMID |
| [104] | Prunet N, Yang WB, Das P, Meyerowitz EM, Jack TP (2017). SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc Natl Acad Sci USA 114, 7166-7171. |
| [105] |
Quon TL, Lampugnani ER, Smyth DR (2017). PETAL LOSS and ROXY1 interact to limit growth within and between sepals but to promote petal initiation in Arabidopsis thaliana. Front Plant Sci 8, 152.
DOI PMID |
| [106] |
Reyes JC, Hennig L, Gruissem W (2002). Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol 130, 1090-1101.
PMID |
| [107] | Rodriguez K, Perales M, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV (2016). DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc Natl Acad Sci USA 113, E6307-E6315. |
| [108] | Running MP, Meyerowitz EM (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261-1269. |
| [109] | Sakai H, Medrano LJ, Meyerowitz EM (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199-203. |
| [110] | Salamah A, Prihatiningsih R, Rostina I, Dwiranti A (2018). Comparative morphology of single and double flowers in Hibiscus rosa-sinensis L. (Malvaceae): a homeosis study. AIP Conf Proc 2023, 020136. |
| [111] |
Sasaki K, Yamaguchi H, Nakayama M, Aida R, Ohtsubo N (2014). Co-modification of class B genes TfDEF and TfGLO in Torenia fournieri Lind. alters both flower morphology and inflorescence architecture. Plant Mol Biol 86, 319-334.
DOI PMID |
| [112] | Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635-644. |
| [113] | Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS (2007). The ABC model and its applicability to basal angiosperms. Ann Bot 100, 155-163. |
| [114] |
Song SS, Qi TC, Huang H, Ren QC, Wu DW, Chang CQ, Peng W, Liu YL, Peng JR, Xie DL (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000-1013.
DOI URL |
| [115] |
Spencer V, Kim M (2018). Re“CYC”ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol 79, 16-26.
DOI URL |
| [116] | Sui MJ, Yan HJ, Wang ZZ, Qiu XQ, Jian HY, Wang QG, Chen M, Zhang H, Tang KX (2019). Identification of microRNA associated with flower organ development in Rosa chinensis ‘Viridiflora’. Plant Sci J 37, 37-46. (in Chinese) |
| 眭梦洁, 晏慧君, 王珍珍, 邱显钦, 蹇洪英, 王其刚, 陈敏, 张颢, 唐开学 (2019). 月季‘绿萼’花器官发育相关microRNA的鉴定及分析. 植物科学学报 37, 37-46. | |
| [117] |
Sun B, Xu YF, Ng KH, Ito T (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23, 1791-1804.
DOI URL |
| [118] | Sun FH, Fang HY, Wen XH, Zhang LS (2023). Phylogenetic and expression analysis of MADS-box gene family in Rhododendron ovatum. Chin Bull Bot 58, 404-416. (in Chinese) |
| 孙福辉, 方慧仪, 温小蕙, 张亮生 (2023). 马银花MADS-box基因家族系统进化与表达分析. 植物学报 58, 404-416. | |
| [119] |
Sun KR, Xue YQ, Prijic Z, Wang SL, Markovic T, Tian CH, Wang YY, Xue JQ, Zhang XX (2022). DNA demethylation induces tree peony flowering with a low deformity rate compared to gibberellin by inducing PsFT expression under forcing culture conditions. Int J Mol Sci 23, 6632.
DOI URL |
| [120] |
Sun QW, Zhou DX (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105, 13679-13684.
DOI PMID |
| [121] | Sun YK, Li JY, Yin HF, Fan ZQ, Zhou XW (2013). Cloning and expression analysis of C function CjAGL6 gene cDNA from Camellia japonica ‘Jinpanlizhi’. Bull Bot Res 33, 330-338. (in Chinese) |
|
孙迎坤, 李纪元, 殷恒福, 范正琪, 周兴文 (2013). 重瓣山茶花‘金盘荔枝’ C功能基因CjAGL6的全长克隆与表达分析. 植物研究 33, 330-338.
DOI |
|
| [122] |
Tähtiharju S, Rijpkema AS, Vetterli A, Albert VA, Teeri TH, Elomaa P (2012). Evolution and diversification of the CYC/TB1 gene family in Asteraceae—a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol Biol Evol 29, 1155-1166.
DOI PMID |
| [123] | Takeda S, Matsumoto N, Okada K (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425-434. |
| [124] | Tang RH, Zhang JC, Zhuang WJ, Wu WR (2003). Progress of the SUPERMAN epigenetic mutation in Arabidopsis. Hereditas 25, 620-622. (in Chinese) |
| 唐荣华, 张君诚, 庄伟建, 吴为人 (2003). 拟南芥SUPERMAN基因表观突变的研究进展. 遗传 25, 620-622. | |
| [125] | Theißen G (2001). Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4, 75-85. |
| [126] | Theißen G, Melzer R, Rümpler F (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259-3271. |
| [127] | Theißen G, Saedler H (2001). Floral quartets. Nature 409, 469-471. |
| [128] | Thomson B, Wellmer F (2019). Molecular regulation of flower development. Curr Top Dev Biol 131, 185-210. |
| [129] | Thomson B, Zheng BB, Wellmer F (2017). Floral organogenesis: when knowing your ABCs is not enough. Plant Physiol 173, 56-64. |
| [130] | van Tunen AJ, Eikelboom W, Angenent GC (1993). Floral organogenesis in Tulipa. Flowering Newsl (16), 33-35, 37-38. |
| [131] |
Varga-Weisz P (2001). ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20, 3076-3085.
PMID |
| [132] |
Wade PA (2001). Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene 20, 3166-3173.
PMID |
| [133] | Wang JD, Zhou Y, Yu JW, Fan XL, Zhang CQ, Li QF, Liu QQ (2020). Advances in the regulation of plant growth and development and stress response by miR172-AP2 module. Chin Bull Bot 55, 205-215. (in Chinese) |
|
王劲东, 周豫, 余佳雯, 范晓磊, 张昌泉, 李钱峰, 刘巧泉 (2020). miR172-AP2模块调控植物生长发育及逆境响应的研究进展. 植物学报 55, 205-215.
DOI |
|
| [134] |
Wellmer F, Graciet E, Riechmann JL (2014). Specification of floral organs in Arabidopsis. J Exp Bot 65, 1-9.
DOI PMID |
| [135] | Wen XH (2019). The construction of genetic network underlying ray and disc florets on capitulum in Chrysanthemum lavandulifolium. Doctoral dissertation. Beijing: Beijing Forestry University. pp. 69-92. (in Chinese) |
| 温小蕙 (2019). 甘菊头状花序上舌状花和管状花发育调控网络的构建. 博士论文. 北京: 北京林业大学. pp. 69-92. | |
| [136] |
Wen XH, Qi S, Huang H, Wu XY, Zhang BH, Fan GX, Yang LW, Hong Y, Dai SL (2019). The expression and interactions of ABCE-class and CYC2-like genes in the capitulum development of Chrysanthemum lavandulifolium and C. × morifolium. Plant Growth Regul 88, 205-214.
DOI |
| [137] |
Wu F, Shi XW, Lin XL, Liu Y, Chong K, Theißen G, Meng Z (2017). The ABCs of flower development: mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J 89, 310-324.
DOI URL |
| [138] |
Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li ZT, Su YH, Slewinski TL, Wagner D (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109, 3576-3581.
DOI URL |
| [139] | Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109, 13452-13457. |
| [140] |
Xia Y, Shi M, Chen WW, Hu RQ, Jing DL, Wu D, Wang SM, Li QF, Deng HH, Guo QG, Liang GL (2020). Expression pattern and functional characterization of PISTILLATA ortholog associated with the formation of petaloid sepals in double-flower Eriobotrya japonica (Rosaceae). Front Plant Sci 10, 1685.
DOI URL |
| [141] | Xiong YY, Wang J (2019). Review of the research progress and prospect of double petals. Nor Hortic (9), 153-158. (in Chinese) |
| 熊阳阳, 王锦 (2019). 重瓣花研究进展与展望. 北方园艺 (9), 153-158. | |
| [142] |
Xu F, Zhang KL, Grunstein M (2005). Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375-385.
PMID |
| [143] |
Xu YF, Prunet N, Gan ES, Wang YB, Stewart D, Wellmer F, Huang JB, Yamaguchi N, Tatsumi Y, Kojima M, Kiba T, Sakakibara H, Jack TP, Meyerowitz EM, Ito T (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J 37, e97499.
DOI URL |
| [144] | Yadav RK, Girke T, Pasala S, Xie MT, Reddy GV (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106, 4941-4946. |
| [145] |
Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25, 2025-2030.
DOI URL |
| [146] |
Yan HJ, Shi SC, Ma N, Cao XQ, Zhang H, Qiu XQ, Wang QG, Jian HY, Zhou NN, Zhang Z, Tang KX (2018). Graft-accelerated virus-induced gene silencing facilitates functional genomics in rose flowers. J Integr Plant Biol 60, 34-44.
DOI |
| [147] | Zahn LM, Kong HZ, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209-2223. |
| [148] |
Zaret KS, Mango SE (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37, 76-81.
DOI PMID |
| [149] |
Zhang CL, Wei LD, Yu XM, Li H, Wang WJ, Wu SZ, Duan F, Bao MZ, Chan Z, He YH (2021). Functional conservation and divergence of SEPALLATA-like genes in the development of two-type florets in marigold. Plant Sci 309, 110938.
DOI URL |
| [150] |
Zhang DY, Zhao XW, Li YY, Ke SJ, Yin WL, Lan SR, Liu ZJ (2022). Advances and prospects of orchid research and industrialization. Hortic Res 9, uhac220.
DOI URL |
| [151] | Zhang JH, Zhang SG, Qi LW, Tong ZK (2014). Research advances in post-transcriptional modification and degradation of mature microRNAs in plants. Chin Bull Bot 49, 483-489. (in Chinese) |
|
张俊红, 张守攻, 齐力旺, 童再康 (2014). 植物成熟microRNA转录后修饰与降解的研究进展. 植物学报 49, 483-489.
DOI |
|
| [152] | Zhang K, Guo XX, Liu XG, Guo L (2018). Advances in research on floral meristem determinacy mechanisms in plants. Chin J Eco-Agricul 26, 1573-1584. (in Chinese) |
| 张科, 郭鑫鑫, 刘西岗, 郭琳 (2018). 植物花分生组织终止发育机制的研究进展. 中国生态农业学报 26, 1573-1584. | |
| [153] | Zhang LW, Eugeni EE, Parthun MR, Freitas MA (2003). Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112, 77-86. |
| [154] | Zhang T, Zhao YF, Juntheikki I, Mouhu K, Broholm SK, Rijpkema AS, Kins L, Lan TY, Albert VA, Teeri TH, Elomaa P (2017). Dissecting functions of SEPALLATA- like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol 216, 939-954. |
| [155] | Zhao YF, Broholm SK, Wang F, Rijpkema AS, Lan TY, Albert VA, Teeri TH, Elomaa P (2020). TCP and MADS- box transcription factor networks regulate heteromorphic flower type identity in Gerbera hybrida. Plant Physiol 184, 1455-1468. |
| [156] | Zhao YQ, Liu QL (2009). Research advances in the formation mechanism and genetic characters of double flowers. Acta Bot Boreal Occident Sin 29, 832-841. (in Chinese) |
| 赵印泉, 刘青林 (2009). 重瓣花的形成机理及遗传特性研究进展. 西北植物学报 29, 832-841. | |
| [157] |
Zhao YX, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16, 2586-2600.
DOI PMID |
| [158] |
Zheng GH, Wei W, Li YP, Kan LJ, Wang FX, Zhang X, Li F, Liu ZC, Kang CY (2019). Conserved and novel roles of miR164-CUC2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytol 224, 480-492.
DOI PMID |
| [1] | Chen Pengxiang, Wang Bo, Wang Zijun, Han Rong. The Regulatory Roles of the Transcription Factors in Plant's Response to UV-B Radiation [J]. Chinese Bulletin of Botany, 2025, 60(3): 449-459. |
| [2] | Xiaoping Lian, Getachew Melaku, Shilai Zhang, Fengyi Hu. MADS-box Genes Driven Life History Strategy Diversity in Brassicaceae [J]. Chinese Bulletin of Botany, 2024, 59(3): 351-354. |
| [3] | Xinhai Zeng, Rui Chen, Yu Shi, Chaoyue Gai, Kai Fan, Zhaowei Li. Research Advances in Biological Functions of Plant SPL Transcription Factors [J]. Chinese Bulletin of Botany, 2023, 58(6): 982-997. |
| [4] | Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang. Phylogenetic and Expression Analysis of MADS-box Gene Family in Rhododendron ovatum [J]. Chinese Bulletin of Botany, 2023, 58(3): 404-416. |
| [5] | Yu Miao, Ruan Chengjiang, Ding Jian, Li Jingbin, Lu Shunguang, Wen Xiufeng. Hrh-miRn458 Regulates Oil Biosynthesis of Sea Buckthorn via Targeting Transcription Factor WRI1 [J]. Chinese Bulletin of Botany, 2022, 57(5): 635-648. |
| [6] | Liu Ting, Wang Tianhao, Chun Yan, Li Xueyong, Zhao Jinfeng. Research Progresses on Epigenetic Regulation of Plant Branching/Tillering [J]. Chinese Bulletin of Botany, 2022, 57(4): 532-548. |
| [7] | Jingwen Wang, Xingjun Wang, Changle Ma, Pengcheng Li. A Review on the Mechanism of Ribosome Stress Response in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 80-89. |
| [8] | Tianxingzi Wang, Zheng Zhu, Yue Chen, Yuqing Liu, Gaowei Yan, Shan Xu, Tong Zhang, Jinjiao Ma, Shijuan Dou, Liyun Li, Guozhen Liu. Rice OsWRKY42 is a Novel Element in Xa21-mediated Resistance Pathway Against Bacterial Leaf Blight [J]. Chinese Bulletin of Botany, 2021, 56(6): 687-698. |
| [9] | Dong Liu. Managing Both Internal and Foreign Affairs—A PHR-centered Gene Network Regulates Plant-mycorrhizal Symbiosis [J]. Chinese Bulletin of Botany, 2021, 56(6): 647-650. |
| [10] | Kaicheng Kang, Xiqiang Niu, Xianzhong Huang, Nengbing Hu, Yihu Sui, Kaijing Zhang, Hao Ai. Genome-wide Identification and Comparative Evolutionary Analysis of the R2R3-MYB Transcription Factor Gene Family in Pepper [J]. Chinese Bulletin of Botany, 2021, 56(3): 315-329. |
| [11] | Yan Wang, Bowei Jia, Mingzhe Sun, Xiaoli Sun. Advances in Molecular Mechanisms of Stress Tolerance in Wild Soybean [J]. Chinese Bulletin of Botany, 2021, 56(1): 104-115. |
| [12] | Ruifeng Yao,Daoxin Xie. New Insight into Strigolactone Signaling [J]. Chinese Bulletin of Botany, 2020, 55(4): 397-402. |
| [13] | Liwen Yang,Shuangrong Liu,Yuhong Li,Rongcheng Lin. Methods for Examining Transcription Factor-DNA Interaction in Plants [J]. Chinese Bulletin of Botany, 2020, 55(4): 468-474. |
| [14] | Lin Hong,Lei Yang,Haijian Yang,Wu Wang. Research Advances in AP2/ERF Transcription Factors in Regulating Plant Responses to Abiotic Stress [J]. Chinese Bulletin of Botany, 2020, 55(4): 481-496. |
| [15] | Yu Zhang, Mingjie Zhao, Wei Zhang. Transcriptional Regulatory Network of Secondary Cell Wall Biosynthesis in Plants [J]. Chinese Bulletin of Botany, 2020, 55(3): 351-368. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||