

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (3): 404-416.DOI: 10.11983/CBB22105 cstr: 32102.14.CBB22105
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang( )
)
Received:2022-05-21
Accepted:2022-08-24
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: zls83@zju.edu.cn
Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang. Phylogenetic and Expression Analysis of MADS-box Gene Family in Rhododendron ovatum[J]. Chinese Bulletin of Botany, 2023, 58(3): 404-416.
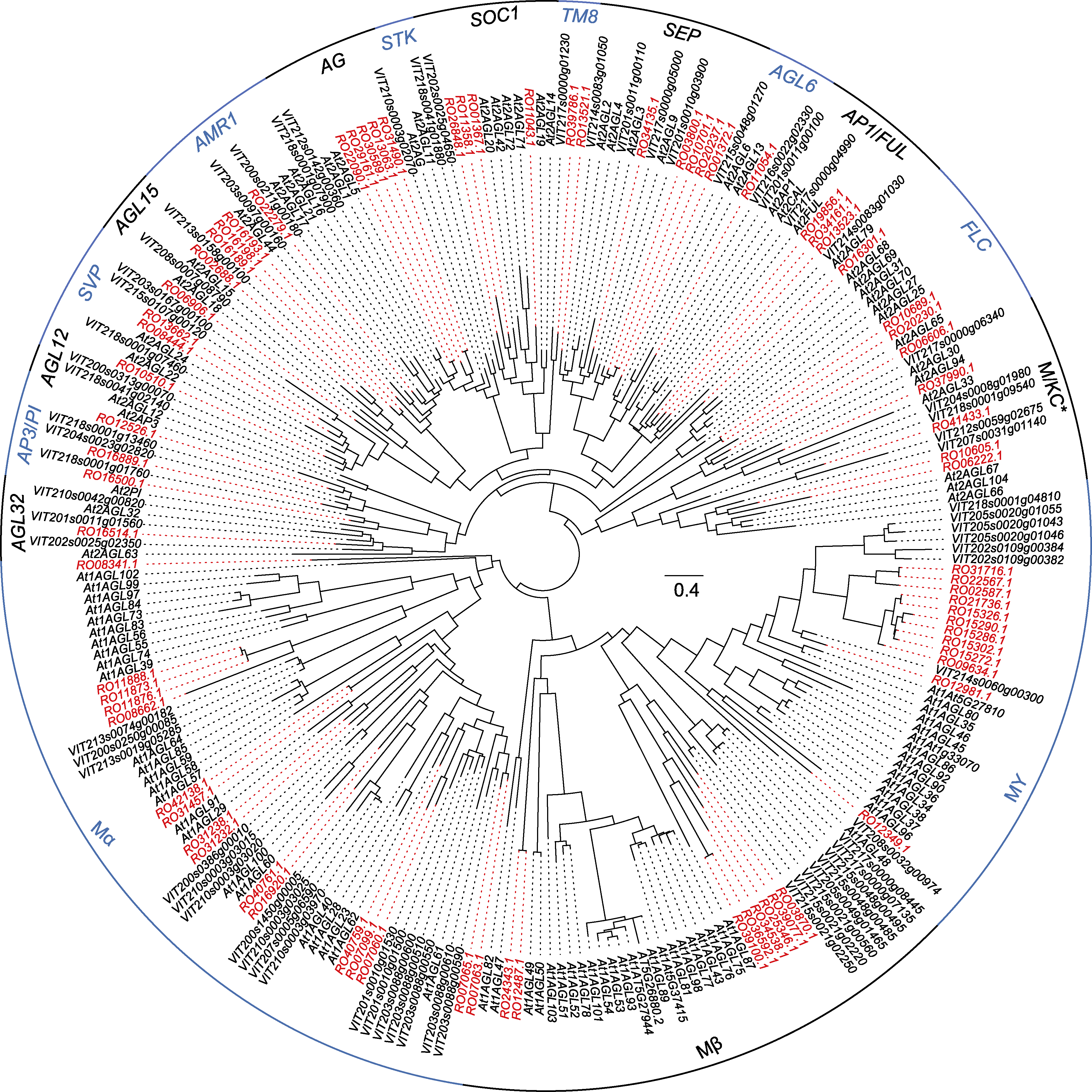
Figure 2 Maximum likelihood phylogenetic tree constructed by Rhododendron ovatum, Arabidopsis thaliana and Vitis vinifera MADS-box genes R. ovatum homologous genes are in red font.
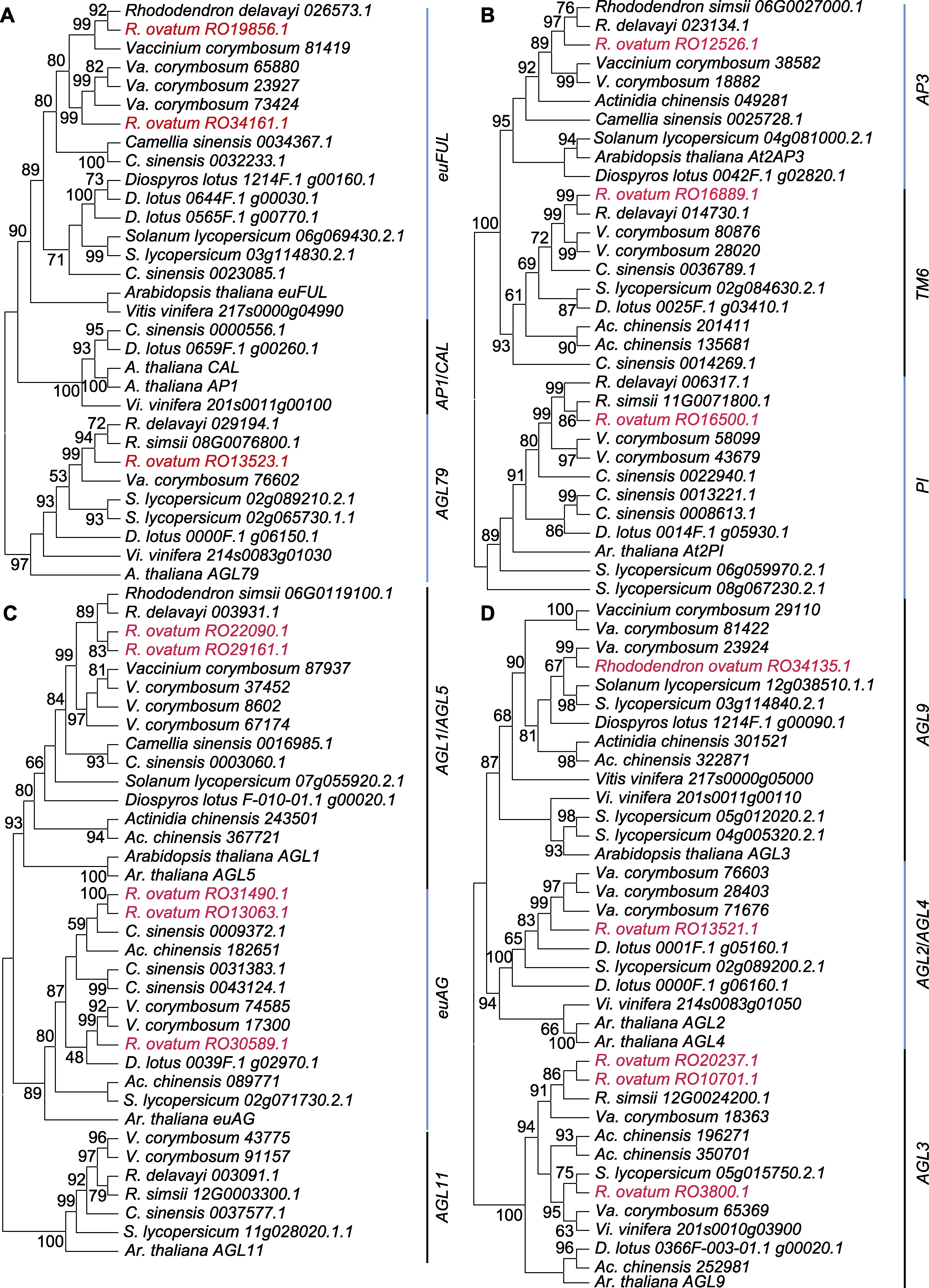
Figure 3 Maximum likelihood phylogenetic trees of floral identity genes (A) Phylogenetic tree of AP1/FUL genes; (B) Phylogenetic tree of AP3/PI genes; (C) Phylogenetic tree of AG genes; (D) Phylogenetic tree of SEP genes. Rhododendron ovatum homologous genes are in red font.
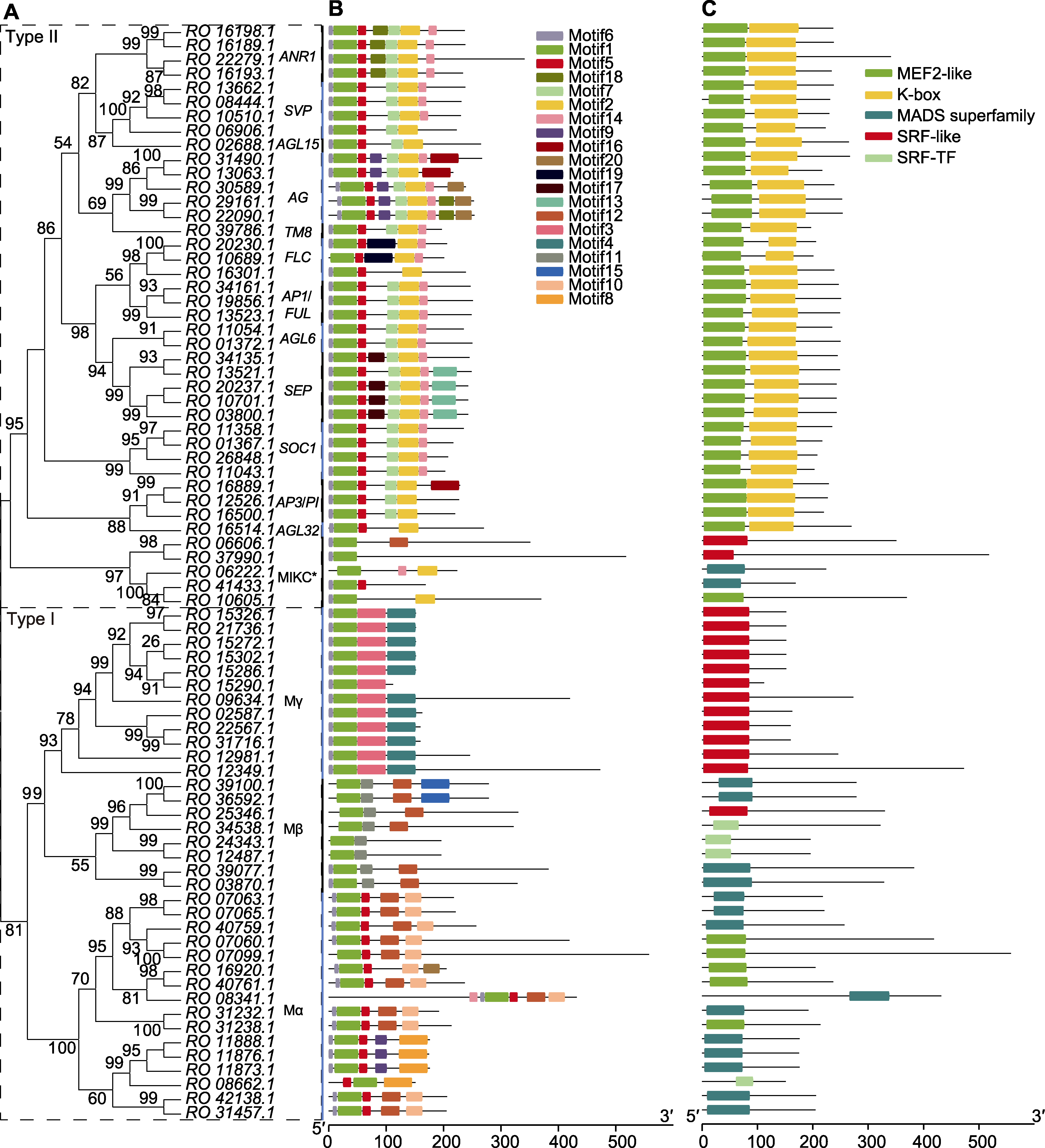
Figure 4 Analysis of structure of Rhododendron ovatum MADS-box protein (A) Phylogenetic tree of R. ovatum MADS-box genes; (B) Conserved motif analysis; (C) Conserved domain analysis
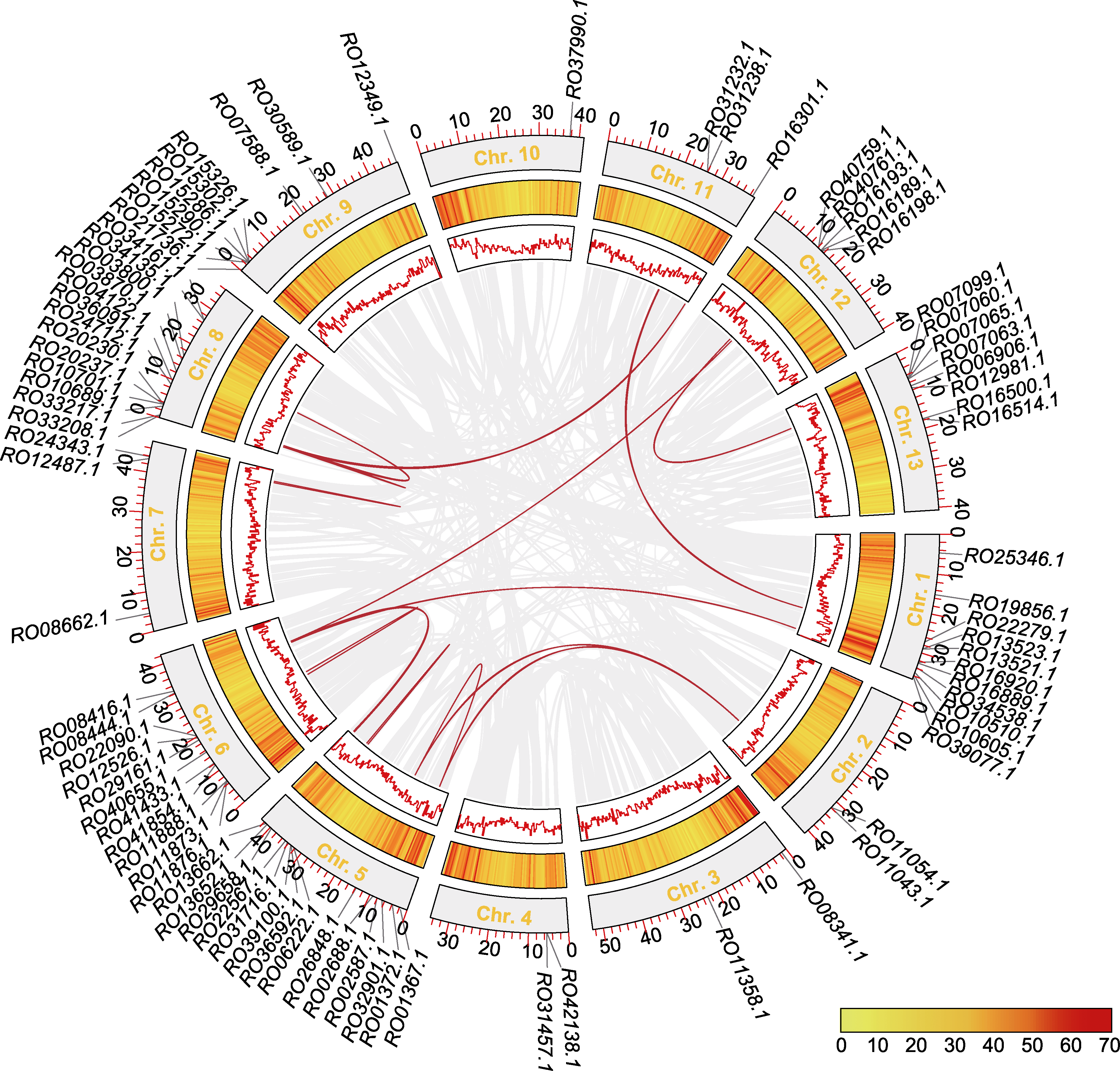
Figure 6 Chromosomal localization and collinearity analysis of Rhododendron ovatum MADS-box genes The tracks from outer to inner circles represent the gene location on chromosomes, gene density, and collinear regions (red lines indicate MADS-box genes).
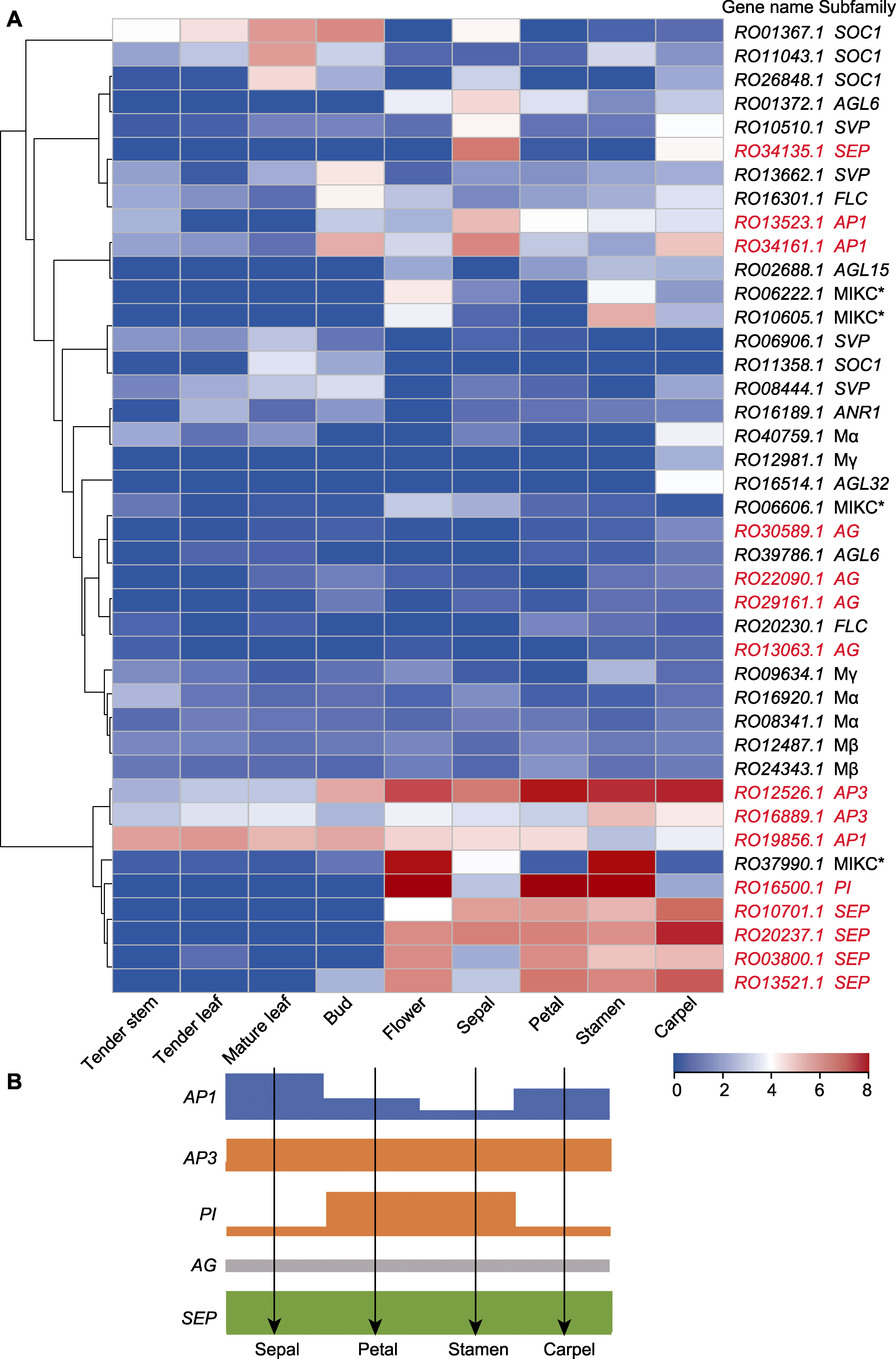
Figure 7 Expression analysis of Rhododendron ovatum MADS-box genes (A) Expression analysis of R. ovatum MADS-box genes in different tissues (the floral identity genes are in red); (B) Proposed ABCE flower development model of R. ovatum (the bar height represents the expression level)
| [1] | 丁献华, 陈伟, 张直峰 (2020). 荷花B类MADS-box基因家族NnDEF基因的克隆及表达分析. 上海农业学报 36, 1-7. |
| [2] |
高虎虎, 张云霄, 胡胜武, 郭媛 (2017). 甘蓝型油菜MADS- box基因家族的鉴定与系统进化分析. 植物学报 52, 699-712.
DOI |
| [3] |
高玮林, 张力曼, 薛超玲, 张垚, 刘孟军, 赵锦 (2022). 枣E类MADS基因在花和果中的表达及其蛋白互作研究. 园艺学报 49, 739-748.
DOI |
| [4] | 耿兴敏, 宦智群, 苏家乐, 刘晓青 (2021). 杜鹃花属植物种质创新研究进展. 分子植物育种 19, 604-613. |
| [5] | 景丹龙, 郭启高, 陈薇薇, 夏燕, 吴頔, 党江波, 何桥, 梁国鲁 (2018). 被子植物花器官发育的模型演变和分子调控. 植物生理学报 54, 355-362. |
| [6] |
景丹龙, 夏燕, 张守攻, 王军辉 (2016). 黄金树B类MADS- box基因表达特征分析. 植物学报 51, 210-217.
DOI |
| [7] |
王莹, 穆艳霞, 王锦 (2021). MADS-box基因家族调控植物花器官发育研究进展. 浙江农业学报 33, 1149-1158.
DOI |
| [8] | 徐启江, 关录飞, 吴笑女, 孙丽, 谭文勃, 聂玉哲, 李玉花 (2008). 草原龙胆MADS-box基因的克隆及表达分析. 植物学通报 25, 415-429. |
| [9] | 杨华, 宋绪忠 (2016). 高温胁迫对马银花的生理指标影响. 林业科技通讯 01, 3-6. |
| [10] | 杨丽, 严露露, 李慧娥, 杜致辉, 郑淇元 (2021). 映山红杜鹃MADS基因家族的鉴定与分析. 分子植物育种 19, 6290-6301. |
| [11] |
Airoldi CA, Davies B (2012). Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genomics 39, 157-165.
DOI PMID |
| [12] |
Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, De Pouplana LR, Martínez-Castilla L, Yanofsky MF (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97, 5328-5333.
PMID |
| [13] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI PMID |
| [14] |
Chen F, Zhang XT, Liu X, Zhang LS (2017). Evolutionary analysis of MIKCC-type MADS-box genes in gymnosperms and angiosperms. Front Plant Sci 8, 895.
DOI PMID |
| [15] |
Duarte JM, Cui LY, Wall PK, Zhang Q, Zhang XH, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006). Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23, 469-478.
PMID |
| [16] |
Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792-1797.
DOI PMID |
| [17] |
Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628-632.
DOI PMID |
| [18] |
Litt A, Kramer EM (2010). The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol 21, 129-137.
DOI PMID |
| [19] |
Liu Y, Cui SJ, Wu F, Yan S, Lin XL, Du XQ, Chong K, Schilling S, Theißen G, Meng Z (2013). Functional conservation of MIKC*-type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288-1303.
DOI URL |
| [20] |
Pařenicová L, De Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538-1551.
DOI URL |
| [21] |
Price MN, Dehal PS, Arkin AP (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641-1650.
DOI PMID |
| [22] | Qiu YC, Köhler C (2022). Endosperm evolution by duplicated and neofunctionalized Type I MADS-box transcription factors. Mol Biol Evol 39, msab355. |
| [23] |
Ruelens P, Zhang ZC, Van Mourik H, Maere S, Kaufmann K, Geuten K (2017). The origin of floral organ identity quartets. Plant Cell 29, 229-242.
DOI URL |
| [24] |
Shan HY, Cheng J, Zhang R, Yao X, Kong HZ (2019). Developmental mechanisms involved in the diversification of flowers. Nat Plants 5, 917-923.
DOI PMID |
| [25] |
Smaczniak C, Immink RGH, Angenent GC, Kaufmann K (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081-3098.
DOI PMID |
| [26] |
Theißen G, Kim JT, Saedler H (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43, 484-516.
PMID |
| [27] |
Theißen G, Melzer R, Rümpler F (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259-3271.
DOI PMID |
| [28] |
Vekemans D, Proost S, Vanneste K, Coenen H, Viaene T, Ruelens P, Maere S, Van De Peer Y, Geuten K (2012). Gamma paleohexaploidy in the stem lineage of core eudicots: significance for MADS-box gene and species diversification. Mol Biol Evol 29, 3793-3806.
DOI PMID |
| [29] |
Wang XY, Gao Y, Wu XP, Wen XH, Li DQ, Zhou H, Li Z, Liu B, Wei JF, Chen F, Chen F, Zhang CJ, Zhang LS, Xia YP (2021). High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol J 19, 2544-2560.
DOI PMID |
| [30] | Wen XH, Li JZ, Wang LL, Lu CF, Gao Q, Xu P, Pu Y, Zhang QL, Hong Y, Hong L, Huang H, Xin HG, Wu XY, Kang DR, Gao K, Li YJ, Ma CF, Li XM, Zheng HK, Wang ZC, Jiao YN, Zhang LS, Dai SL (2022). The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic Res 9, uhab022. |
| [31] |
Wu WW, Huang XT, Cheng J, Li ZG, De Folter S, Huang ZR, Jiang XQ, Pang HX, Tao SH (2011). Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Mol Biol Evol 28, 501-511.
DOI PMID |
| [32] |
Yang FS, Nie S, Liu H, Shi TL, Tian XC, Zhou SS, Bao YT, Jia KH, Guo JF, Zhao W, An N, Zhang RG, Yun QZ, Wang XZ, Mannapperuma C, Porth I, El-Kassaby YA, Street NR, Wang XR, Van De Peer Y, Mao JF (2020). Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat Commun 11, 5269.
DOI |
| [33] |
Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Lohaus R, Chang XJ, Dong W, Ho SYW, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, Van De Peer Y, Tang HB (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79-84.
DOI |
| [1] | Jie Cao, Qiulian Lu, Jianping Zhai, Baohui Liu, Chao Fang, Shichen Li, Tong Su. Changes in the Expression of the Soybean TPS Gene Family Under Salt Stress and Haplotype Selection Analysis [J]. Chinese Bulletin of Botany, 2025, 60(2): 172-185. |
| [2] | Qiaoxia Li, Youlong Li, Jigang Li, Chenlong Chen, Kun Sun. Effects of photoperiods on the development of chasmogamous and cleistogamous flowers in Viola monbeigii and V. dissecta [J]. Biodiv Sci, 2024, 32(6): 23484-. |
| [3] | Yegeng Fan,Lihang Qiu,Xing Huang,Huiwen Zhou,Chongkun Gan,Yangrui Li,Rongzhong Yang,Jianming Wu,Rongfa Chen. Expression Analysis of Key Genes in Gibberellin Biosynthesis and Related Phytohormonal Dynamics During Sugarcane Internode Elongation [J]. Chinese Bulletin of Botany, 2019, 54(4): 486-496. |
| [4] | Li Ma, Wancang Sun, Jinhai Yuan, Zigang Liu, Junyan Wu, Yan Fang, Yaozhao Xu, Yuanyuan Pu, Jing Bai, Xiaoyun Dong, Huili He. Expression Analysis of β-1,3-Glucanase Gene from Winter Brassica rapa Under Low Temperature Stress [J]. Chinese Bulletin of Botany, 2017, 52(5): 568-578. |
| [5] | Baoling Liu, Li Zhang, Yan Sun,Jinai Xue, Changyong Gao, Lixia Yuan, Jiping Wang, Xiaoyun Jia, Runzhi Li. Genome-wide Characterization of bZIP Transcription Factors in Foxtail Millet and Their Expression Profiles in Response to Drought and Salt Stresses [J]. Chinese Bulletin of Botany, 2016, 51(4): 473-487. |
| [6] | Danlong Jing, Yan Xia, Shougong Zhang, Junhui Wang. Expression Analysis of B-class MADS-box Genes from Catalpa speciosa [J]. Chinese Bulletin of Botany, 2016, 51(2): 210-217. |
| [7] | Yuanbao Cai, Xiangyan Yang, Guangming Sun, Qiang Huang, Yeqiang Liu, Shaopeng Li, Zhili Zhang. Cloning of Flowering-related Gene AcMADS1 and Characterization of Expression in Tissues of Pineapple (Ananas comosus) [J]. Chinese Bulletin of Botany, 2014, 49(6): 692-703. |
| [8] | Ying Zhai, Xiaojie Yang, Tianguo Sun, Yan Zhao, Chunfen Yu, Xiuwen Wang. Cloning, Expressing and Functional Analysis of GmERF5 from Soybean [J]. Chinese Bulletin of Botany, 2013, 48(5): 498-506. |
| [9] | Danlong Jing, Jiang Ma, Bo Zhang, Yiyang Han, Zhixiong Liu, Faju Chen. Expression Analysis of MwAG in Different Organs and Developmental Stages of Magnolia wufengensis [J]. Chinese Bulletin of Botany, 2013, 48(2): 145-150. |
| [10] | Junhong Zhang, Shougong Zhang, Tao Wu, Suying Han, Wenhua Yang, Liwang Qi. Expression Analysis of Five MiRNAs and Their Precursors During Somatic Embryogenesis in Larch [J]. Chinese Bulletin of Botany, 2012, 47(5): 462-473. |
| [11] | Huajun Guo;Yuannian Jiao;Chao Di;Dongxia Yao;Gaihua Zhang;Xue Zheng;Lan Liu;Qunlian Zhang;Aiguang Guo*;Zhen Su*. Discovery of Arabidopsis GRAS Family Genes in Response to Osmotic and Drought Stresses [J]. Chinese Bulletin of Botany, 2009, 44(03): 290-299. |
| [12] | Qijiang Xu;Lufei Guan;Xiaonü Wu;Li Sun;Wenbo Tan;Yuzhe Nie;Yuhua Li. Cloning and Expression Analysis of MADS-box Genes from Lisianthus (Eustoma grandiflorum) [J]. Chinese Bulletin of Botany, 2008, 25(04): 415-429. |
| [13] | LU Lu WANG Hong LI De-Zhu. Advances in Molecular Phylogenetics and Biogeography of Gaultheria (Ericaceae) [J]. Chinese Bulletin of Botany, 2005, 22(06): 658-667. |
| [14] | MA Yue-Ping CHEN Fan DAI Si-Lan. Studies of LEAFY Homologue Genes in Higher Plants [J]. Chinese Bulletin of Botany, 2005, 22(05): 605-613. |
| [15] | ZHU Gen-Fa GUO Zhen-Fei. Progress on Molecular Biology of Main Ornamental Orchidaceae [J]. Chinese Bulletin of Botany, 2004, 21(04): 471-477. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||