

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (1): 150-158.DOI: 10.11983/CBB22108 cstr: 32102.14.CBB22108
Special Issue: 杂粮生物学专辑 (2023年58卷1期)
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Weijun Ye, Yin Zhang, Peiran Wang, Lingling Zhang, Dongfeng Tian, Zejiang Wu, Bin Zhou*( )
)
Received:2022-05-25
Accepted:2022-07-25
Online:2023-01-01
Published:2023-01-05
Contact:
*E-mail: 18756019871@139.com
Weijun Ye, Yin Zhang, Peiran Wang, Lingling Zhang, Dongfeng Tian, Zejiang Wu, Bin Zhou. QTLs Analysis for Five Yield-related Traits in Mungbean[J]. Chinese Bulletin of Botany, 2023, 58(1): 150-158.
| Year | Traits | Parental lines | F2 or F3 population | |||||
|---|---|---|---|---|---|---|---|---|
| Weilv11 | Sulv16-10 | Means±SD | Range | CV (%) | Skewness | Kurtosis | ||
| 2019 (F2) | NPP | 11.0 | 20.2 ** | 18.26±8.86 | 5.0-46.0 | 48.52 | 1.27 | 1.26 |
| NSP | 10.1 | 12.0 ** | 10.62±0.98 | 9.6-14.3 | 7.77 | -0.84 | 0.55 | |
| HSW (g) | 5.00 | 7.42 ** | 6.33±0.69 | 4.77-8.19 | 10.95 | 0.27 | -0.45 | |
| BMS | 0.5 | 3.0 ** | 2.08±1.48 | 0.0-6.0 | 71.17 | 0.35 | -0.45 | |
| YP (g) | 5.21 | 14.95 ** | 11.48±5.95 | 2.67-28.96 | 51.80 | 1.16 | 0.82 | |
| 2020 (F3) | NPP | 9.4 | 23.2 ** | 16.94±8.65 | 5.0-48.0 | 51.05 | 1.38 | 2.01 |
| NSP | 9.8 | 12.3 ** | 10.41±1.30 | 8.5-15.0 | 10.15 | -0.73 | 0.80 | |
| HSW (g) | 5.06 | 7.02 ** | 5.47±0.68 | 3.40-7.20 | 12.51 | -0.44 | 0.27 | |
| BMS | 0.5 | 3.6 ** | 1.71±1.39 | 0.0-6.0 | 81.68 | -0.11 | -1.17 | |
| YP (g) | 4.62 | 16.89 ** | 8.94±4.71 | 2.00-5.40 | 52.70 | 1.15 | 1.18 | |
Table 1 Phenotype analysis for the mungbean parental lines, F2 and F3 populations
| Year | Traits | Parental lines | F2 or F3 population | |||||
|---|---|---|---|---|---|---|---|---|
| Weilv11 | Sulv16-10 | Means±SD | Range | CV (%) | Skewness | Kurtosis | ||
| 2019 (F2) | NPP | 11.0 | 20.2 ** | 18.26±8.86 | 5.0-46.0 | 48.52 | 1.27 | 1.26 |
| NSP | 10.1 | 12.0 ** | 10.62±0.98 | 9.6-14.3 | 7.77 | -0.84 | 0.55 | |
| HSW (g) | 5.00 | 7.42 ** | 6.33±0.69 | 4.77-8.19 | 10.95 | 0.27 | -0.45 | |
| BMS | 0.5 | 3.0 ** | 2.08±1.48 | 0.0-6.0 | 71.17 | 0.35 | -0.45 | |
| YP (g) | 5.21 | 14.95 ** | 11.48±5.95 | 2.67-28.96 | 51.80 | 1.16 | 0.82 | |
| 2020 (F3) | NPP | 9.4 | 23.2 ** | 16.94±8.65 | 5.0-48.0 | 51.05 | 1.38 | 2.01 |
| NSP | 9.8 | 12.3 ** | 10.41±1.30 | 8.5-15.0 | 10.15 | -0.73 | 0.80 | |
| HSW (g) | 5.06 | 7.02 ** | 5.47±0.68 | 3.40-7.20 | 12.51 | -0.44 | 0.27 | |
| BMS | 0.5 | 3.6 ** | 1.71±1.39 | 0.0-6.0 | 81.68 | -0.11 | -1.17 | |
| YP (g) | 4.62 | 16.89 ** | 8.94±4.71 | 2.00-5.40 | 52.70 | 1.15 | 1.18 | |
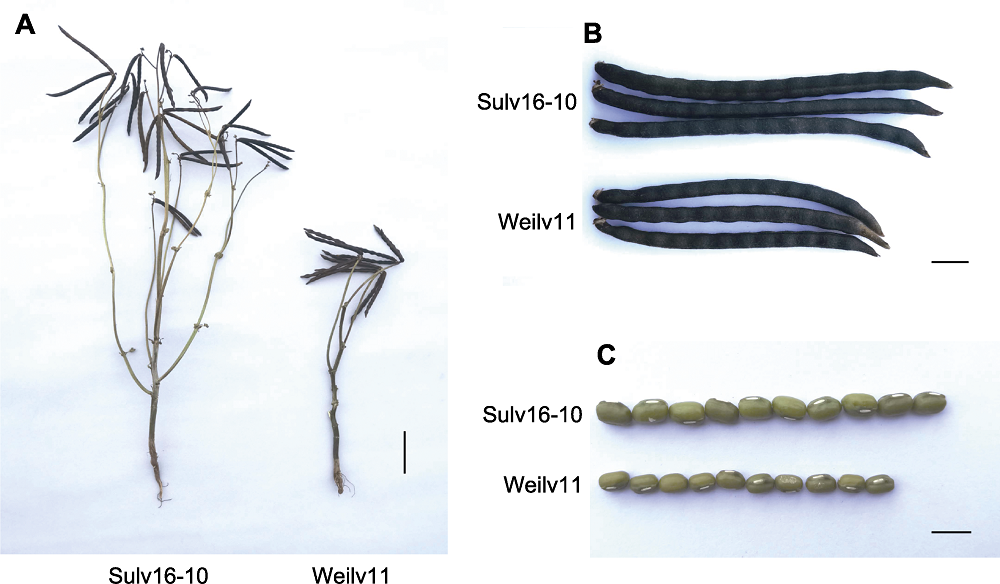
Figure 1 Phenotype of mungbean parental lines (A) Phenotype of parental plants at mature stage (bar=5 cm); (B) Phenotype of the mature pods of parental lines (bar=1 cm); (C) Phenotype of parental seeds (bar=5 mm)
| Traits | NPP | NSP | HSW | BMS |
|---|---|---|---|---|
| NSP | 0.362 ** | |||
| 0.357 ** | ||||
| HSW | -0.005 | -0.049 | ||
| 0.149 | 0.125 | |||
| BMS | 0.403 ** | 0.190 * | 0.174 * | |
| 0.708 ** | 0.423 ** | 0.311 ** | ||
| YP | 0.950 ** | 0.409 ** | 0.163 | 0.425 ** |
| 0.914 ** | 0.464 ** | 0.318 ** | 0.701 ** |
Table 2 Correlation analysis of yield-related agronomic traits in mungbean
| Traits | NPP | NSP | HSW | BMS |
|---|---|---|---|---|
| NSP | 0.362 ** | |||
| 0.357 ** | ||||
| HSW | -0.005 | -0.049 | ||
| 0.149 | 0.125 | |||
| BMS | 0.403 ** | 0.190 * | 0.174 * | |
| 0.708 ** | 0.423 ** | 0.311 ** | ||
| YP | 0.950 ** | 0.409 ** | 0.163 | 0.425 ** |
| 0.914 ** | 0.464 ** | 0.318 ** | 0.701 ** |
| Year | Traits | QTL name | Chromosome | Marker interval | Position (cM) | Likelihood of odd (LOD) | Add effect | Phenotype variance explained (%) |
|---|---|---|---|---|---|---|---|---|
| 2019 | NPP | qNPP3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 3.47 | 4.62 | 11.09 |
| NSP | qNSP3 | 3 | ID3-5-ID3-6 | 13.16-23.40 | 5.92 | 0.58 | 17.93 | |
| HSW | qHSW3 | 3 | ID3-7-ID3-9 | 29.12-31.34 | 3.78 | 0.26 | 5.33 | |
| qHSW7 | 7 | ID7-10-ID7-7 | 100.12-103.50 | 23.15 | 0.71 | 46.07 | ||
| qHSW10 | 10 | ID10-3-ID10-4 | 6.84-9.08 | 2.90 | -0.09 | 4.24 | ||
| BMS | qBMS3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 6.44 | 1.03 | 18.51 | |
| qBMS11 | 11 | ID11-6-ID11-7 | 33.46-35.32 | 2.85 | 0.25 | 7.06 | ||
| YP | qYP3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 4.60 | 3.67 | 14.18 | |
| 2020 | NPP | qNPP3a | 3 | ID3-4-ID3-5 | 9.83-13.06 | 3.39 | 3.63 | 2.27 |
| NSP | qNSP3a | 3 | R3-9-R3-12 | 20.88-22.23 | 6.37 | 0.66 | 19.93 | |
| HSW | qHSW7a | 7 | ID7-10-ID7-7 | 29.77-31.76 | 8.80 | 0.42 | 30.17 |
Table 3 QTL analysis for yield-related agronomic traits in mungbean
| Year | Traits | QTL name | Chromosome | Marker interval | Position (cM) | Likelihood of odd (LOD) | Add effect | Phenotype variance explained (%) |
|---|---|---|---|---|---|---|---|---|
| 2019 | NPP | qNPP3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 3.47 | 4.62 | 11.09 |
| NSP | qNSP3 | 3 | ID3-5-ID3-6 | 13.16-23.40 | 5.92 | 0.58 | 17.93 | |
| HSW | qHSW3 | 3 | ID3-7-ID3-9 | 29.12-31.34 | 3.78 | 0.26 | 5.33 | |
| qHSW7 | 7 | ID7-10-ID7-7 | 100.12-103.50 | 23.15 | 0.71 | 46.07 | ||
| qHSW10 | 10 | ID10-3-ID10-4 | 6.84-9.08 | 2.90 | -0.09 | 4.24 | ||
| BMS | qBMS3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 6.44 | 1.03 | 18.51 | |
| qBMS11 | 11 | ID11-6-ID11-7 | 33.46-35.32 | 2.85 | 0.25 | 7.06 | ||
| YP | qYP3 | 3 | ID3-8-ID3-9 | 30.60-31.34 | 4.60 | 3.67 | 14.18 | |
| 2020 | NPP | qNPP3a | 3 | ID3-4-ID3-5 | 9.83-13.06 | 3.39 | 3.63 | 2.27 |
| NSP | qNSP3a | 3 | R3-9-R3-12 | 20.88-22.23 | 6.37 | 0.66 | 19.93 | |
| HSW | qHSW7a | 7 | ID7-10-ID7-7 | 29.77-31.76 | 8.80 | 0.42 | 30.17 |
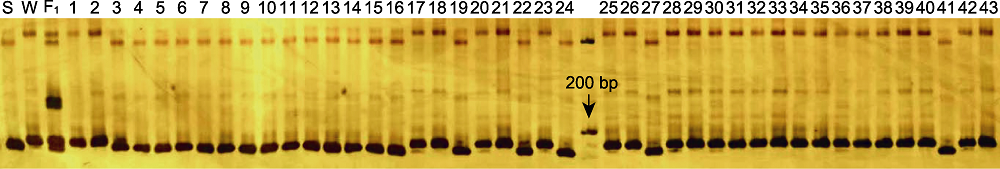
Figure 3 Genotype analysis for mungbean accessions using marker R7-13.4 S: Sulv16-10; W: Weilv11. 1-24 are accessions with large seed size; 25-43 are accessions with small seed size.
| No. | Variety name | HSW (g) | Genotype | No. | Variety name | HSW (g) | Genotype |
|---|---|---|---|---|---|---|---|
| 1 | Baolv200520 | 7.13±0.21 | aa | 23 | Jilv0816 | 6.63±0.32 | aa |
| 2 | E1002 | 7.53±0.40 | aa | 24 | Sulv15-11 | 6.67±0.31 | AA |
| 3 | E1006 | 6.80±0.30 | AA | 25 | Bailv11 | 4.60±0.10 | aa |
| 4 | E1007 | 6.90±0.26 | AA | 26 | E1003 | 4.70±0.20 | aa |
| 5 | Elv3 | 7.13±0.31 | AA | 27 | Jinlv4 | 4.87±0.23 | AA |
| 6 | Jilv7 | 7.03±0.21 | AA | 28 | Jinlv6 | 4.30±0.10 | aa |
| 7 | Su2074 | 7.90±0.56 | AA | 29 | Lulv1002-3 | 4.83±0.15 | aa |
| 8 | Sukang4 | 7.03±0.23 | AA | 30 | Pinlv08116 | 4.67±0.15 | aa |
| 9 | Sukang1 | 7.37±0.25 | AA | 31 | Su09-8 | 4.37±0.31 | aa |
| 10 | Sulv016 | 7.13±0.31 | AA | 32 | Suhei2 | 4.30±0.00 | aa |
| 11 | Sulv023 | 7.07±0.35 | AA | 33 | Suheilv | 4.23±0.06 | aa |
| 12 | Sulv11-8 | 7.23±0.15 | AA | 34 | Sukang2 | 5.00±0.10 | aa |
| 13 | Sulv203 | 8.20±0.26 | AA | 35 | Taiyuan52 | 3.70±0.20 | aa |
| 14 | Sulv209 | 7.07±0.32 | AA | 36 | Taiyuanchuanfu | 4.73±0.15 | aa |
| 15 | Sulv4 | 8.23±0.64 | AA | 37 | Taiyuanzao2 | 5.00±0.36 | aa |
| 16 | Sulv9073 | 6.97±0.25 | AA | 38 | Tong1188326 | 3.80±0.26 | aa |
| 17 | Zhonglv1 | 7.27±0.35 | aa | 39 | Wei2117 | 4.60±0.44 | aa |
| 18 | Zhonglv3 | 6.90±0.44 | aa | 40 | Weilv12 | 4.80±0.35 | aa |
| 19 | Zhonglv5 | 6.87±0.45 | AA | 41 | Weilv2117 | 4.57±0.42 | AA |
| 20 | Baolv200810 | 6.53±0.58 | aa | 42 | Weilv8 | 4.93±0.23 | aa |
| 21 | E1009 | 6.63±0.40 | aa | 43 | Yinggelv | 4.57±0.15 | aa |
| 22 | E75-3 | 6.77±0.40 | AA |
Table 4 Phenotype and genotype of 43 mungbean accessions
| No. | Variety name | HSW (g) | Genotype | No. | Variety name | HSW (g) | Genotype |
|---|---|---|---|---|---|---|---|
| 1 | Baolv200520 | 7.13±0.21 | aa | 23 | Jilv0816 | 6.63±0.32 | aa |
| 2 | E1002 | 7.53±0.40 | aa | 24 | Sulv15-11 | 6.67±0.31 | AA |
| 3 | E1006 | 6.80±0.30 | AA | 25 | Bailv11 | 4.60±0.10 | aa |
| 4 | E1007 | 6.90±0.26 | AA | 26 | E1003 | 4.70±0.20 | aa |
| 5 | Elv3 | 7.13±0.31 | AA | 27 | Jinlv4 | 4.87±0.23 | AA |
| 6 | Jilv7 | 7.03±0.21 | AA | 28 | Jinlv6 | 4.30±0.10 | aa |
| 7 | Su2074 | 7.90±0.56 | AA | 29 | Lulv1002-3 | 4.83±0.15 | aa |
| 8 | Sukang4 | 7.03±0.23 | AA | 30 | Pinlv08116 | 4.67±0.15 | aa |
| 9 | Sukang1 | 7.37±0.25 | AA | 31 | Su09-8 | 4.37±0.31 | aa |
| 10 | Sulv016 | 7.13±0.31 | AA | 32 | Suhei2 | 4.30±0.00 | aa |
| 11 | Sulv023 | 7.07±0.35 | AA | 33 | Suheilv | 4.23±0.06 | aa |
| 12 | Sulv11-8 | 7.23±0.15 | AA | 34 | Sukang2 | 5.00±0.10 | aa |
| 13 | Sulv203 | 8.20±0.26 | AA | 35 | Taiyuan52 | 3.70±0.20 | aa |
| 14 | Sulv209 | 7.07±0.32 | AA | 36 | Taiyuanchuanfu | 4.73±0.15 | aa |
| 15 | Sulv4 | 8.23±0.64 | AA | 37 | Taiyuanzao2 | 5.00±0.36 | aa |
| 16 | Sulv9073 | 6.97±0.25 | AA | 38 | Tong1188326 | 3.80±0.26 | aa |
| 17 | Zhonglv1 | 7.27±0.35 | aa | 39 | Wei2117 | 4.60±0.44 | aa |
| 18 | Zhonglv3 | 6.90±0.44 | aa | 40 | Weilv12 | 4.80±0.35 | aa |
| 19 | Zhonglv5 | 6.87±0.45 | AA | 41 | Weilv2117 | 4.57±0.42 | AA |
| 20 | Baolv200810 | 6.53±0.58 | aa | 42 | Weilv8 | 4.93±0.23 | aa |
| 21 | E1009 | 6.63±0.40 | aa | 43 | Yinggelv | 4.57±0.15 | aa |
| 22 | E75-3 | 6.77±0.40 | AA |
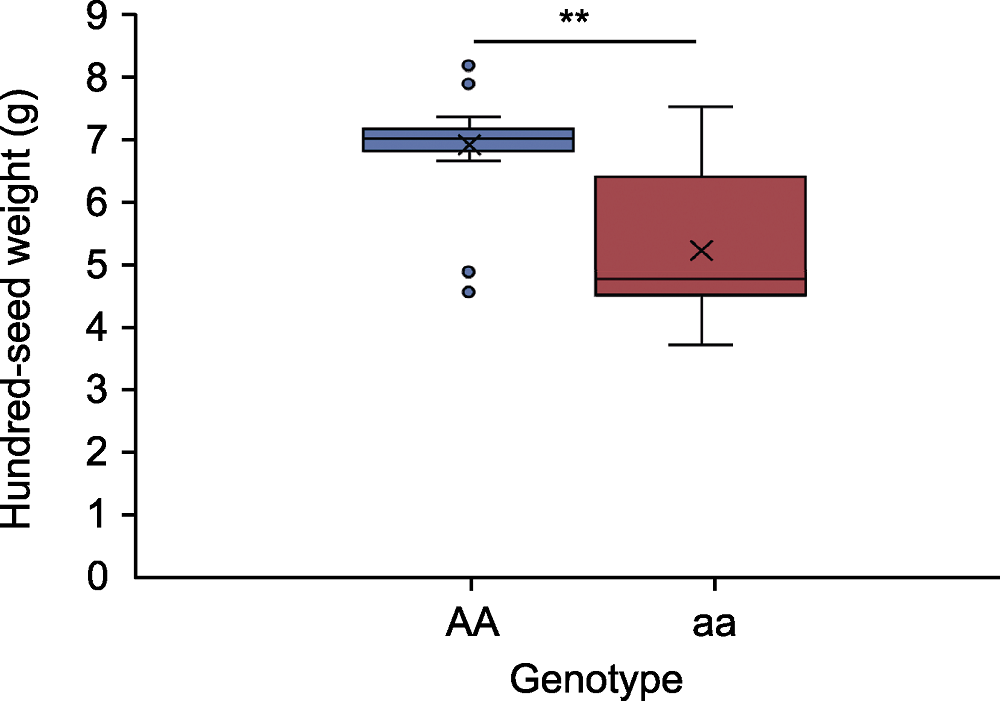
Figure 4 Statistical analysis of genotype and phenotype for 43 mungbean accessions AA and aa represent Sulv16-10 and Weilv11 genotype, respectively. ** indicates extremely significant difference (P<0.01).
| [1] | 陈红霖, 胡亮亮, 杨勇, 郝曦煜, 李姝彤, 王素华, 王丽侠, 程须珍 (2020). 481份国内外绿豆种质农艺性状及豆象抗性鉴定评价及遗传多样性分析. 植物遗传资源学报 21, 549-559. |
| [2] |
陈吉宝 (2020). 绿豆产量性状的QTL定位. 中国农业科技导报 22(10), 38-48.
DOI |
| [3] | 程须珍, 王素华, 王丽霞 (2006). 绿豆种质资源描述规范和数据标准. 北京: 中国农业出版社. pp. 9-22. |
| [4] | 杜梦柯, 连文婷, 张晓, 李欣欣 (2021). 氮处理对大豆根瘤固氮能力及GmLbs基因表达的影响. 植物学报 56, 391-403. |
| [5] |
何恒斌, 贾桂霞 (2013). 豆科植物早期共生信号转导的研究进展. 植物学报 48, 665-675.
DOI |
| [6] | 侯小峰, 刘静, 王彩萍, 左联忠, 赵吉平, 郭鹏燕, 郭兆萍 (2015). 绿豆产量与主要农艺性状的灰色关联分析. 作物杂志 (1), 53-56. |
| [7] |
马秀杰 (2014). 间作对绿豆生物性状、产量和品质的影响. 核农学报 28, 546-551.
DOI |
| [8] | 梅丽, 程须珍, 王素华, 王丽侠, 蔡庆生, 刘春吉, 徐宁, 刘长友, 孙蕾 (2011). 绿豆产量相关农艺性状的QTL定位. 植物遗传资源学报 12, 948-956. |
| [9] | 王健康 (2009). 数量性状基因的完备区间作图方法. 作物学报 35, 239-245. |
| [10] | 王洁 (2020). 绿豆高密度遗传图谱构建及重要农艺性状QTL定位. 硕士论文. 北京: 中国农业科学院. pp. 22-26. |
| [11] | 徐东旭 (2015). 冀西北绿豆产量与主要农艺性状的灰色关联度分析. 农业科技通讯 (7), 96-98, 251. |
| [12] | 杨春玲, 王阔, 关立, 侯军红, 宋志均, 韩勇, 葛鹏飞 (2005). 绿豆主要农艺性状间的相关及通径分析. 杂粮作物 25, 314-315. |
| [13] | 杨芳, 杨媛, 冯高, 杨明君 (2012). 绿豆籽粒产量与主要农艺性状的相关分析. 农业科技通讯 (7), 95-97. |
| [14] | 杨勇, 周斌, 杨超华, 张丽亚 (2015). 夏播绿豆不同品种产量与主要农艺性状的相关分析. 作物杂志 (4), 65-68. |
| [15] | Alam MK, Islam MM, Salahin N, Hasanuzzaman M (2014). Effect of tillage practices on soil properties and crop productivity in wheat-mungbean-rice cropping system under subtropical climatic conditions. Sci World J 2014, 437283. |
| [16] |
Chankaew S, Somta P, Sorajjapinun W, Srinives P (2011). Quantitative trait loci mapping of Cercospora leaf spot resistance in mungbean, Vigna radiata (L.) Wilczek. Mol Breeding 28, 255-264.
DOI URL |
| [17] | Chen HM, Ku HM, Schafleitner R, Bains TS, Kuo CG, Liu CA, Nair RM (2013). The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 192, 205-216. |
| [18] |
Chotechung S, Somta P, Chen JB, Yimram T, Chen X, Srinives P (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: bruchidae) resistance in mungbean (Vigna radiata). Theor Appl Genet 129, 1673-1683.
DOI PMID |
| [19] |
Fatokun CA, Menancio-Hautea DI, Danesh D, Young ND (1992). Evidence for orthologous seed weight genes in cowpea and mungbean based on RFLP mapping. Genetics 132, 841-846.
DOI PMID |
| [20] | Ha J, Satyawan D, Jeong H, Lee E, Cho KH, Kim MY, Lee SH (2021). A near-complete genome sequence of mungbean (Vigna radiata L.) provides key insights into the modern breeding program. Plant Genome 14, e20121. |
| [21] |
Humphry ME, Lambrides CJ, Chapman SC, Aitken EAB, Imrie BC, Lawn RJ, Mcintyre CL, Liu CJ (2005). Relationships between hard-seededness and seed weight in mungbean (Vigna radiata) assessed by QTL analysis. Plant Breeding 124, 292-298.
DOI URL |
| [22] |
Humphry ME, Magner T, McIntyre CL, Aitken EAB, Liu CJ (2003). Identification of a major locus conferring resistance to powdery mildew (Erysiphe polygoni DC) in mungbean (Vigna radiata L. Wilczek) by QTL analysis. Genome 46, 738-744.
DOI PMID |
| [23] |
Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 7, e41304.
DOI URL |
| [24] | Kajonphol T, Sangsiri C, Somta P, Toojinda T, Srinives P (2012). SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). SABRAO J Breed Genet 44, 71-86. |
| [25] |
Kang YJ, Kim SK, Kim MY, Lestari P, Kim KH, Ha BK, Jun TH, Hwang WJ, Lee T, Lee J, Shim S, Yoon MY, Jang YE, Han KS, Taeprayoon P, Yoon N, Somta P, Tanya P, Kim KS, Gwag JG, Moon JK, Lee YH, Park BS, Bombarely A, Doyle JJ, Jackson SA, Schafleitner R, Srinives P, Varshney RK, Lee SH (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun 5, 5443.
DOI |
| [26] |
Keatinge JDH, Easdown WJ, Yang RY, Chadha ML, Shanmugasundaram S (2011). Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica 180, 129-141.
DOI URL |
| [27] |
Kim SK, Nair RM, Lee J, Lee SH (2015). Genomic resources in mungbean for future breeding programs. Front Plant Sci 6, 626.
DOI PMID |
| [28] |
Kitsanachandee R, Somta P, Chatchawankanphanich O, Akhtar KP, Shah TM, Nair RM, Bains TS, Sirari A, Kaur L, Srinives P (2013). Detection of quantitative trait loci for mungbean yellow mosaic India virus (MYMIV) resistance in mungbean (Vigna radiata (L.) Wilczek) in India and Pakistan. Breeding Sci 63, 367-373.
DOI PMID |
| [29] |
Mei L, Cheng XZ, Wang SH, Wang LX, Liu CY, Sun L, Xu N, Humphry ME, Lambrides CJ, Li HB, Liu CJ (2009). Relationship between bruchid resistance and seed mass in mungbean based on QTL analysis. Genome 52, 589-596.
DOI PMID |
| [30] |
Sompong U, Somta P, Raboy V, Srinives P (2012). Mapping of quantitative trait loci for phytic acid and phosphorus contents in seed and seedling of mungbean (Vigna radiata (L.) Wilczed). Breed Sci 62, 87-92.
DOI URL |
| [31] | Somta P, Chankaew S, Kongjaimun A, Srinives P (2015). QTLs controlling seed weight and days to flowering in mungbean [Vigna radiata (L.) Wilczek], their conservation in azuki bean [V. angularis (Ohwi) Ohwi & Ohashi] and rice bean [V. umbellata (Thunb.) Ohwi & Ohashi]. Agrivita 37, 159-168. |
| [32] |
Tuberosa R, Salvi S, Sanguineti MC, Landi P, Maccaferri M, Conti S (2002). Mapping QTLs regulating morpho- physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann Bot 89, 941-963.
DOI URL |
| [33] | Yaqub M, Mahmood T, Akhtar M, Iqbal MM, Ali S (2010). Induction of mungbean [Vigna radiata (L.) Wilczek] as a grain legume in the annual rice-wheat double cropping system. Pak J Bot 42, 3125-3135. |
| [34] |
Ye WJ, Yang Y, Wang PR, Zhang Y, Zhang LY, Tian DF, Zhang L, Zhang LL, Zhou B (2021). InDel marker development and QTL analysis of agronomic traits in mung- bean [Vigna radiate (L.) Wilczek]. Mol Breeding 41, 66.
DOI |
| [35] |
Yundaeng C, Somta P, Chen JB, Yuan XX, Chankaew S, Chen X (2021). Fine mapping of QTL conferring Cercospora leaf spot disease resistance in mungbean revealed TAF5 as candidate gene for the resistance. Theor Appl Genet 134, 701-714.
DOI |
| [36] |
Zhang MC, Wang DM, Zheng Z, Humphry M, Liu CJ (2008). Development of PCR-based markers for a major locus conferring powdery mildew resistance in mungbean (Vigna radiata). Plant Breed 127, 429-432.
DOI URL |
| [1] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [2] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [3] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [4] | WU Han, BAI Jie, LI Jun-Li, Guli JIAPAER, BAO An-Ming. Study of spatio-temporal variation in fractional vegetation cover and its influencing factors in Xinjiang, China [J]. Chin J Plant Ecol, 2024, 48(1): 41-55. |
| [5] | Qiwei Jia, Qianqian Zhong, Yujia Gu, Tianqi Lu, Wei Li, Shuai Yang, Chaoyu Zhu, Chengxiang Hu, Sanfeng Li, Yuexing Wang, Yuchun Rao. Mapping of QTL for Cell Wall Related Components in Rice Stem and Analysis of Candidate Genes [J]. Chinese Bulletin of Botany, 2023, 58(6): 882-892. |
| [6] | Jiayi Jin, Yiting Luo, Huimin Yang, Tao Lu, Hanfei Ye, Jiyi Xie, Kexin Wang, Qianyu Chen, Yuan Fang, Yuexing Wang, Yuchun Rao. QTL Mapping and Expression Analysis on Candidate Genes Related to Chlorophyll Content in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 394-403. |
| [7] | Xiaomin Feng, Xiang Gao, Huadong Zang, Yuegao Hu, Changzhong Ren, Zhiping Hao, Huiqing Lü, Zhaohai Zeng. Intercropping Effect and Nitrogen Transfer Characteristics of Oat-Mungbean Intercrop [J]. Chinese Bulletin of Botany, 2023, 58(1): 122-131. |
| [8] | Wei Heping, Lu Tao, Jia Qiwei, Deng Fei, Zhu Hao, Qi Zehua, Wang Yuxi, Ye Hanfei, Yin Wenjing, Fang Yuan, Mu Dan, Rao Yuchun. QTL Mapping of Candidate Genes for Heading Date in Rice [J]. Chinese Bulletin of Botany, 2022, 57(5): 588-595. |
| [9] | Hanfei Ye, Wenjing Yin, Yian Guan, Kairu Yang, Qianyu Chen, Shuying Yu, Xudong Zhu, Dedong Xin, Wei Zhang, Yuexing Wang, Yuchun Rao. QTL Mapping and Candidate Gene Analysis of Vitamin E in Rice Grain [J]. Chinese Bulletin of Botany, 2022, 57(2): 157-170. |
| [10] | XU Guang-Lai, LI Ai-Juan, XU Xiao-Hua, YANG Xian-Cheng, YANG Qiang-Qiang. NDVIdynamics and driving climatic factors in the Protected Zones for Ecological Functions in China [J]. Chin J Plant Ecol, 2021, 45(3): 213-223. |
| [11] | Chenyang Pan, Yue Zhang, Han Lin, Qianyu Chen, Kairu Yang, Jiaji Jiang, Mengjia Li, Tao Lu, Kexin Wang, Mei Lu, Sheng Wang, Hanfei Ye, Yuchun Rao, Haitao Hu. QTL Mapping and Candidate Gene Analysis on Rice Leaf Water Potential [J]. Chinese Bulletin of Botany, 2021, 56(3): 275-283. |
| [12] | Chenyang Pan, Hanfei Ye, Weiyong Zhou, Sheng Wang, Mengjia Li, Mei Lu, Sanfeng Li, Xudong Zhu, Yuexing Wang, Yuchun Rao, Gaoxing Dai. QTL Mapping of Candidate Genes Involved in Cd Accumulation in Rice Grain [J]. Chinese Bulletin of Botany, 2021, 56(1): 25-32. |
| [13] | Yilan Zhang, Xue Lin, Yi Wu, Mengjia Li, Shengjie Zhang, Mei Lu, Yuchun Rao, Yuexing Wang. Research Progress on Genetics and Breeding of Rice Roots [J]. Chinese Bulletin of Botany, 2020, 55(3): 382-393. |
| [14] | WANG Li-Hua, XUE Jing-Yue, XIE Yu, WU Yan. Spatial distribution and influencing factors of soil organic carbon among different climate types in Sichuan, China [J]. Chin J Plant Ecol, 2018, 42(3): 297-306. |
| [15] | Jiang-Qun LIU, Ming-Yu YIN, Si-Yu ZUO, Shao-Bing YANG, Tana WUYUN. Phenotypic variations in natural populations of Amygdalus pedunculata [J]. Chin J Plant Ecol, 2017, 41(10): 1091-1102. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||