

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (5): 544-558.DOI: 10.11983/CBB21014 cstr: 32102.14.CBB21014
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Dandan Wu1, Yongkun Chen1,2, Yu Yang1, Chunyan Kong1, Ming Gong1,*( )
)
Received:2021-01-18
Accepted:2021-05-07
Online:2021-09-01
Published:2021-08-31
Contact:
Ming Gong
Dandan Wu, Yongkun Chen, Yu Yang, Chunyan Kong, Ming Gong. Identification of the Cysteine Protease Family and Corresponding miRNAs in Jatropha curcas and Their Response to Chill-hardening[J]. Chinese Bulletin of Botany, 2021, 56(5): 544-558.
| Primer name | Sequence (5′→ 3′) |
|---|---|
| miR1314-y-F | GCCGGTCTCCAATGTTAGG |
| miR1151-y-F | ATCTGGTTGTGGGACCCG |
| miR5156-x-F | GCGAGACTGTGAAACTGCAAA |
| miR6483-y-F | GCGTTGTAGAAATTTTCAGGATCA |
| JcXBCP3L-F | TCTGTGGGTATGGATGGTTCTGC |
| JcXBCP3L-R | ACCCCAGTCAGTACCCCACGA |
| JcRD21B-F | CAACGCTTTAGGAGAGAAGGAA |
| JcRD21B-R | ACGCAACTTGTTCCTCCTGAC |
| JcDEK1-F | TGCTGGGAAATTCTGGTG |
| JcDEK1-R | AGCCGTCAAACCCACCA |
| Reverse primer | GCTGTCAACGATACGCTACGTAACG |
| U6-F | GGAACGATACAGAGAAGATT |
| GAPDH-F | TGAAGGACTGGAGAGGTGGAAGAGC |
| GAPDH-R | ATCAACAGTTGGAACACGGAAAGCC |
Table 1 The related primers for qRT-PCR
| Primer name | Sequence (5′→ 3′) |
|---|---|
| miR1314-y-F | GCCGGTCTCCAATGTTAGG |
| miR1151-y-F | ATCTGGTTGTGGGACCCG |
| miR5156-x-F | GCGAGACTGTGAAACTGCAAA |
| miR6483-y-F | GCGTTGTAGAAATTTTCAGGATCA |
| JcXBCP3L-F | TCTGTGGGTATGGATGGTTCTGC |
| JcXBCP3L-R | ACCCCAGTCAGTACCCCACGA |
| JcRD21B-F | CAACGCTTTAGGAGAGAAGGAA |
| JcRD21B-R | ACGCAACTTGTTCCTCCTGAC |
| JcDEK1-F | TGCTGGGAAATTCTGGTG |
| JcDEK1-R | AGCCGTCAAACCCACCA |
| Reverse primer | GCTGTCAACGATACGCTACGTAACG |
| U6-F | GGAACGATACAGAGAAGATT |
| GAPDH-F | TGAAGGACTGGAGAGGTGGAAGAGC |
| GAPDH-R | ATCAACAGTTGGAACACGGAAAGCC |
| Locus ID | Gene name | Subgroup | Chr. | Size (aa) | Mw (Da) | pI |
|---|---|---|---|---|---|---|
| XM_012209792.1 | JcRD21A | C1A | Chr. 10 | 475 | 52682.16 | 5.39 |
| XM_012221410.1 | JcSAG12H4 | C1A | Chr. 4 | 340 | 37417.89 | 5.13 |
| XM_012226309.1 | JcSAG12H1 | C1A | Chr. 3 | 311 | 34256.46 | 6.89 |
| XM_012216029.1 | JcSAG12H2 | C1A | Chr. 7 | 268 | 28610.75 | 4.66 |
| XM_012216557.1 | JcXBCP3 | C1A | Chr. 4 | 441 | 48817.81 | 6.06 |
| XM_012218796.1 | JcRD21A1 | C1A | Chr. 10 | 358 | 40417.25 | 5.10 |
| XM_012223641.1 | JcALP1 | C1A | Chr. 7 | 347 | 38935.92 | 8.43 |
| XM_012225588.1 | JcRD19B | C1A | Chr. 10 | 409 | 44955.43 | 6.71 |
| XM_012230158.1 | JcCEP2 | C1A | Chr. 7 | 358 | 39943.84 | 6.19 |
| XM_012233098.1 | JcSAG12H5 | C1A | Chr. 9 | 340 | 37592.12 | 5.01 |
| XM_012236390.1 | JcXBCP3L | C1A | Chr. 3 | 524 | 58042.85 | 5.24 |
| XM_012219020.1 | JcXCP1 | C1A | Chr. 5 | 349 | 39054.13 | 5.59 |
| XM_012211812.1 | JcRD19A | C1A | Chr. 2 | 370 | 40564.71 | 5.95 |
| XM_012212907.1 | JcRD21B | C1A | Chr. 4 | 466 | 51785.03 | 5.28 |
| XM_012213199.1 | JcSAG12H3 | C1A | Chr. 2 | 339 | 37598.38 | 5.94 |
| XM_012216569.1 | JcRD19C | C1A | Chr. 4 | 368 | 40923.12 | 5.81 |
| XM_012218200.1 | JcXCP2 | C1A | Chr. 4 | 350 | 39176.15 | 5.40 |
| XM_012221663.1 | JcTHI1 | C1A | Chr. 11 | 358 | 39435.49 | 5.88 |
| XM_012223907.1 | JcRD21C | C1A | Chr. 7 | 366 | 41119.35 | 5.40 |
| XM_012224546.1 | JcCEP1 | C1A | Chr. 5 | 360 | 40142.92 | 5.69 |
| XM_020685390.1 | JcSAG12H6 | C1A | Chr. 3 | 181 | 20779.04 | 9.49 |
| XM_020685389.1 | JcSAG12H7 | C1A | Chr. 3 | 181 | 20779.04 | 9.49 |
| XM_012227664.1 | JcCTB1 | C1A | Chr. 8 | 358 | 39709.10 | 6.07 |
| XM_012225478.1 | JcDEK1 | C2 | Chr. 1 | 2158 | 239010.06 | 6.03 |
| XM_012224060.1 | JcUCH2 | C12 | Chr. 4 | 337 | 38597.28 | 6.43 |
| XM_012214672.1 | JcUCH3 | C12 | Chr. 9 | 236 | 26006.51 | 4.66 |
| XM_012231737.1 | JcVPE2 | C13 | Chr. 9 | 495 | 55104.34 | 5.58 |
| XM_012221936.1 | JcVPE1 | C13 | Chr. 11 | 493 | 54253.23 | 5.93 |
| XM_012232060.1 | JcVPE3 | C13 | Chr. 5 | 485 | 54983.84 | 6.22 |
| XM_012229170.1 | JcAMC4G | C14 | Chr. 5 | 418 | 45953.22 | 5.18 |
| Locus ID | Gene name | Subgroup | Chr. | Size (aa) | Mw (Da) | pI |
| XM_012227353.1 | JcAMC9A | C14 | Chr. 3 | 317 | 35221.20 | 5.84 |
| XM_012213747.1 | JcAMC3B | C14 | Chr. 2 | 374 | 41063.04 | 5.71 |
| XM_012213748.1 | JcAMC1C | C14 | Chr. 2 | 330 | 37646.13 | 8.05 |
| XM_012213889.1 | JcAMC2D | C14 | Chr. 2 | 406 | 45165.20 | 8.65 |
| XM_012213890.1 | JcAMC1E | C14 | Chr. 2 | 335 | 37746.43 | 8.10 |
| XM_012228350.1 | JcAMC1F | C14 | Chr. 8 | 362 | 39681.00 | 6.41 |
| XM_012235144.1 | JcAMC1H | C14 | Chr. 5 | 370 | 40503.80 | 6.34 |
| XM_012230023.1 | JcPCP1 | C15 | Chr. 7 | 217 | 23577.96 | 6.07 |
| XM_012233456.1 | JcPCP2 | C15 | Chr. 6 | 219 | 23969.43 | 6.08 |
Table 2 Physicochemical properties of identified cysteine proteins in Jatropha curcas
| Locus ID | Gene name | Subgroup | Chr. | Size (aa) | Mw (Da) | pI |
|---|---|---|---|---|---|---|
| XM_012209792.1 | JcRD21A | C1A | Chr. 10 | 475 | 52682.16 | 5.39 |
| XM_012221410.1 | JcSAG12H4 | C1A | Chr. 4 | 340 | 37417.89 | 5.13 |
| XM_012226309.1 | JcSAG12H1 | C1A | Chr. 3 | 311 | 34256.46 | 6.89 |
| XM_012216029.1 | JcSAG12H2 | C1A | Chr. 7 | 268 | 28610.75 | 4.66 |
| XM_012216557.1 | JcXBCP3 | C1A | Chr. 4 | 441 | 48817.81 | 6.06 |
| XM_012218796.1 | JcRD21A1 | C1A | Chr. 10 | 358 | 40417.25 | 5.10 |
| XM_012223641.1 | JcALP1 | C1A | Chr. 7 | 347 | 38935.92 | 8.43 |
| XM_012225588.1 | JcRD19B | C1A | Chr. 10 | 409 | 44955.43 | 6.71 |
| XM_012230158.1 | JcCEP2 | C1A | Chr. 7 | 358 | 39943.84 | 6.19 |
| XM_012233098.1 | JcSAG12H5 | C1A | Chr. 9 | 340 | 37592.12 | 5.01 |
| XM_012236390.1 | JcXBCP3L | C1A | Chr. 3 | 524 | 58042.85 | 5.24 |
| XM_012219020.1 | JcXCP1 | C1A | Chr. 5 | 349 | 39054.13 | 5.59 |
| XM_012211812.1 | JcRD19A | C1A | Chr. 2 | 370 | 40564.71 | 5.95 |
| XM_012212907.1 | JcRD21B | C1A | Chr. 4 | 466 | 51785.03 | 5.28 |
| XM_012213199.1 | JcSAG12H3 | C1A | Chr. 2 | 339 | 37598.38 | 5.94 |
| XM_012216569.1 | JcRD19C | C1A | Chr. 4 | 368 | 40923.12 | 5.81 |
| XM_012218200.1 | JcXCP2 | C1A | Chr. 4 | 350 | 39176.15 | 5.40 |
| XM_012221663.1 | JcTHI1 | C1A | Chr. 11 | 358 | 39435.49 | 5.88 |
| XM_012223907.1 | JcRD21C | C1A | Chr. 7 | 366 | 41119.35 | 5.40 |
| XM_012224546.1 | JcCEP1 | C1A | Chr. 5 | 360 | 40142.92 | 5.69 |
| XM_020685390.1 | JcSAG12H6 | C1A | Chr. 3 | 181 | 20779.04 | 9.49 |
| XM_020685389.1 | JcSAG12H7 | C1A | Chr. 3 | 181 | 20779.04 | 9.49 |
| XM_012227664.1 | JcCTB1 | C1A | Chr. 8 | 358 | 39709.10 | 6.07 |
| XM_012225478.1 | JcDEK1 | C2 | Chr. 1 | 2158 | 239010.06 | 6.03 |
| XM_012224060.1 | JcUCH2 | C12 | Chr. 4 | 337 | 38597.28 | 6.43 |
| XM_012214672.1 | JcUCH3 | C12 | Chr. 9 | 236 | 26006.51 | 4.66 |
| XM_012231737.1 | JcVPE2 | C13 | Chr. 9 | 495 | 55104.34 | 5.58 |
| XM_012221936.1 | JcVPE1 | C13 | Chr. 11 | 493 | 54253.23 | 5.93 |
| XM_012232060.1 | JcVPE3 | C13 | Chr. 5 | 485 | 54983.84 | 6.22 |
| XM_012229170.1 | JcAMC4G | C14 | Chr. 5 | 418 | 45953.22 | 5.18 |
| Locus ID | Gene name | Subgroup | Chr. | Size (aa) | Mw (Da) | pI |
| XM_012227353.1 | JcAMC9A | C14 | Chr. 3 | 317 | 35221.20 | 5.84 |
| XM_012213747.1 | JcAMC3B | C14 | Chr. 2 | 374 | 41063.04 | 5.71 |
| XM_012213748.1 | JcAMC1C | C14 | Chr. 2 | 330 | 37646.13 | 8.05 |
| XM_012213889.1 | JcAMC2D | C14 | Chr. 2 | 406 | 45165.20 | 8.65 |
| XM_012213890.1 | JcAMC1E | C14 | Chr. 2 | 335 | 37746.43 | 8.10 |
| XM_012228350.1 | JcAMC1F | C14 | Chr. 8 | 362 | 39681.00 | 6.41 |
| XM_012235144.1 | JcAMC1H | C14 | Chr. 5 | 370 | 40503.80 | 6.34 |
| XM_012230023.1 | JcPCP1 | C15 | Chr. 7 | 217 | 23577.96 | 6.07 |
| XM_012233456.1 | JcPCP2 | C15 | Chr. 6 | 219 | 23969.43 | 6.08 |
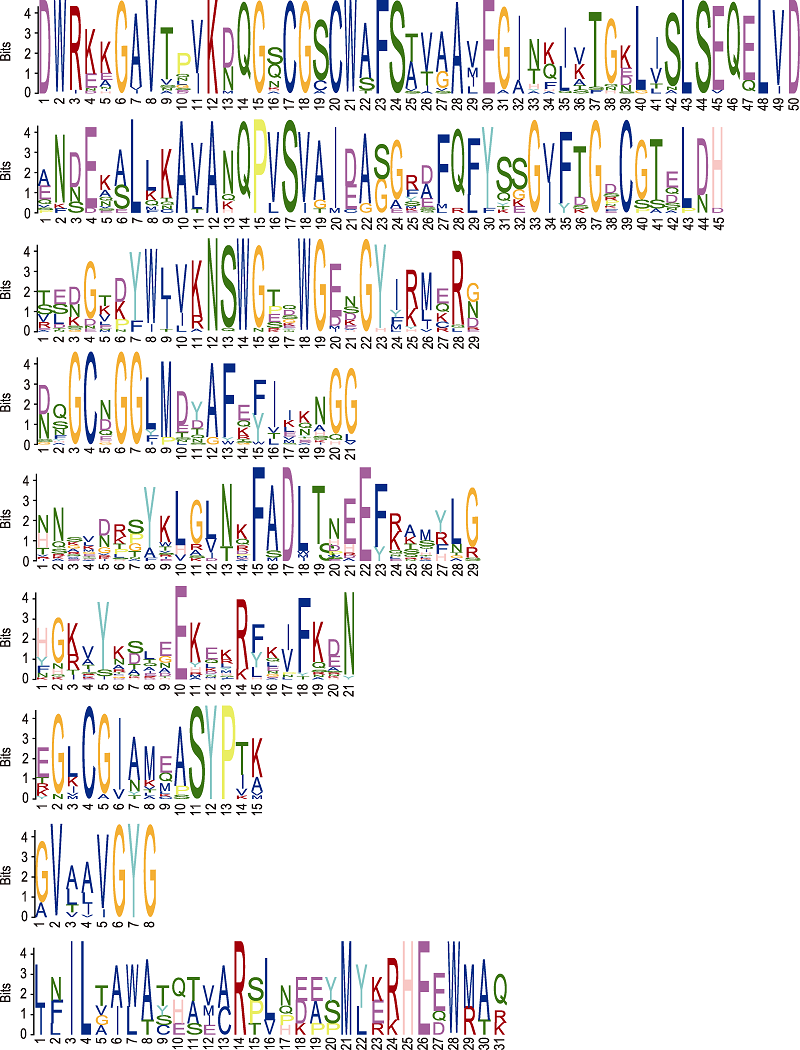
Figure 3 The top nine motifs of cysteine protease family in Jatropha curcas identified using the MEME software The abscissa represents the length of the motif; the ordinate indicates the conservatism of the motif.
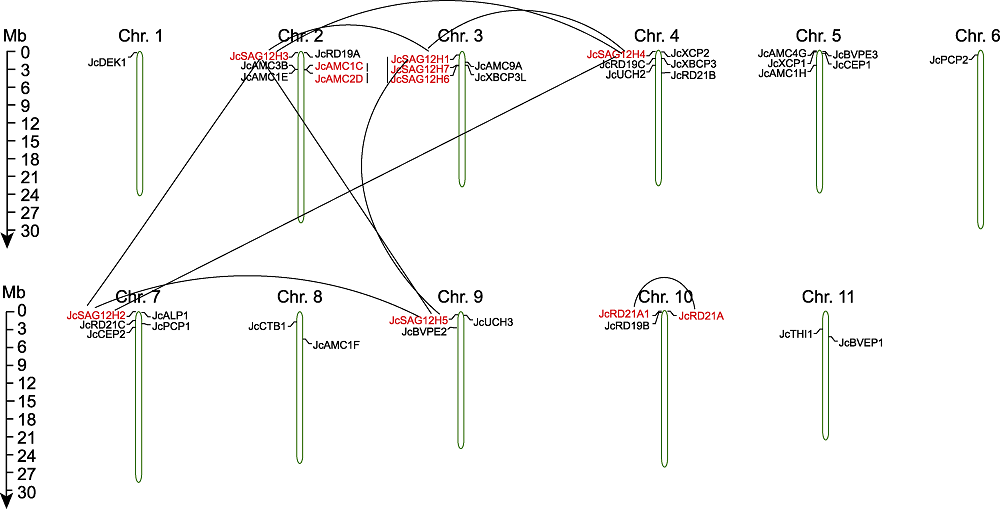
Figure 4 Chromosome location and repetitive events of cysteine protease gene pairs in Jatropha curcas Repeated gene pairs are shown and linked by the lines.
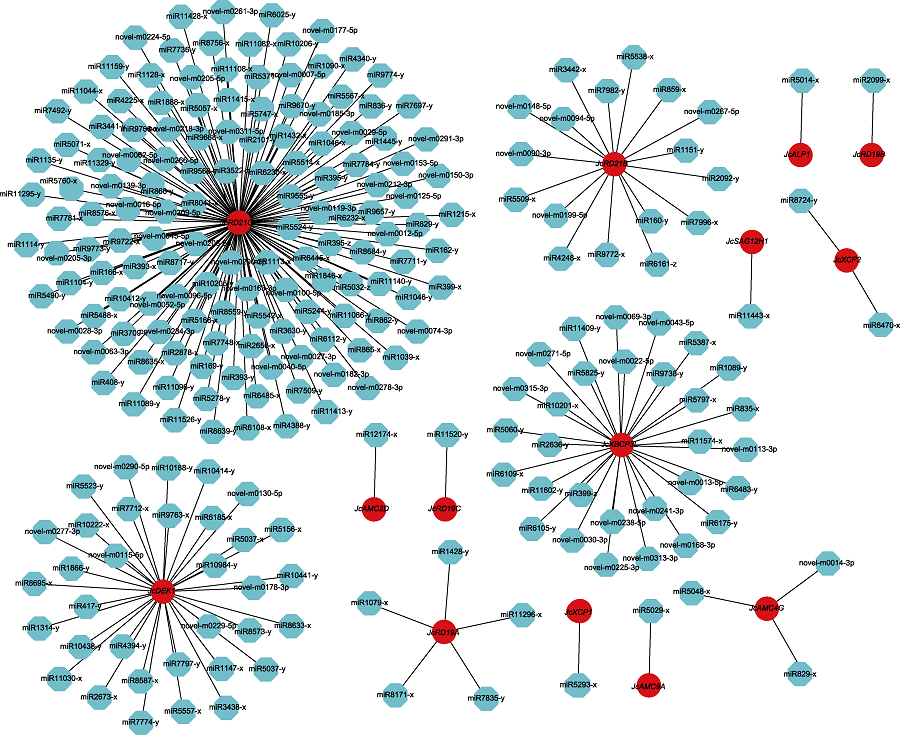
Figure 7 Networks of targeted regulation among miRNAs and their corresponding cysteine protease family genes in Jatropha curcas Red represents cysteine protease gene and blue represents miRNAs.
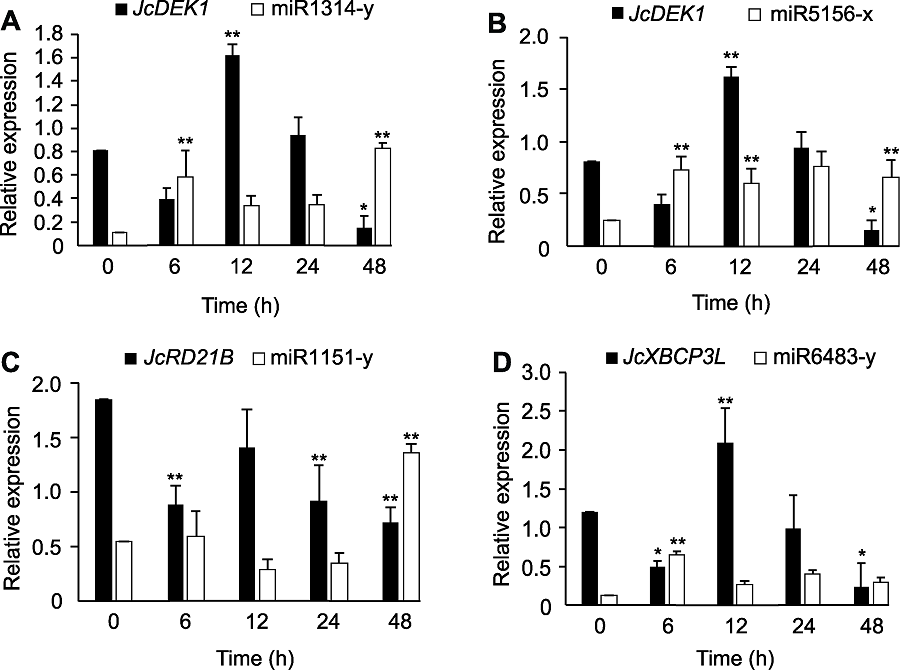
Figure 9 Co-expression analysis by qRT-PCR of the cysteine protease genes JcDEK1, JcRD21B and JcXBCP3L as well as the miRNAs targeting these genes in Jatropha curcas seedlings during chill-hardening at 12°C * and ** indicated the significant differences between the expression level at different time and control 0 h at P<0.05 and P<0.01 level, respectively.
| [1] | 郝大海, 龚明 (2020). miRNA作用机制研究进展. 基因组学与应用生物学 39, 3647-3657. |
| [2] | 孔春艳, 陈永坤, 王莎莎, 郝大海, 杨宇, 龚明 (2019). 小桐子低温胁迫下microRNA实时荧光定量PCR内参的筛选与比较. 生物技术通报 35(7), 25-31. |
| [3] | 李忠光, 龚明 (2011). 不同化学消毒剂对小桐子种子萌发和幼苗生长的影响. 种子 30(2), 4-7, 12. |
| [4] | 宋敏, 张瑶, 王丽莹, 彭向永 (2020). 甘蓝型油菜ZF-HD基因家族的鉴定与系统进化分析. 植物学报 54, 699-710. |
| [5] | 王海波, 郭俊云, 唐利洲 (2019). 小桐子MAPKKKK基因家族的全基因组鉴定及表达分析. 植物生理学报 55, 367-377. |
| [6] |
王劲东, 周豫, 余佳雯, 范晓磊, 张昌泉, 李钱峰, 刘巧泉 (2020). miR172-AP2模块调控植物生长发育及逆境响应的研究进展. 植物学报 55, 205-215.
DOI |
| [7] | 吴丹丹, 陈永坤, 杨宇, 孔春艳, 龚明 (2021). 小桐子cystatin家族基因和相应miRNAs的鉴定及其在低温响应中可能的作用. 植物生理学报 57, 347-361. |
| [8] |
闫晨阳, 陈赢男 (2020). 4种模式植物LRR VIII-2亚家族基因的鉴定和进化历史分析. 植物学报 55, 442-456.
DOI |
| [9] | 张翠桔, 莫蓓莘, 陈雪梅, 崔洁 (2020). 植物miRNA作用方式的分子机制研究进展. 生物技术通报 36(7), 1-14. |
| [10] |
Ahmad R, Zuily-Fodil Y, Passaquet C, Bethenod O, Roche R, Repellin A (2014). Identification and characterization of MOR-CP, a cysteine protease induced by ozone and developmental senescence in maize ( Zea mays L.) leaves. Chemosphere 108, 245-250.
DOI URL |
| [11] |
Ao PX, Li ZG, Fan DM, Gong M (2013a). Involvement of antioxidant defense system in chill hardening-induced chilling tolerance in Jatropha curcas seedlings. Acta Physiol Plant 35, 153-160.
DOI URL |
| [12] |
Ao PX, Li ZG, Gong M (2013b). Involvement of compatible solutes in chill hardening-induced chilling tolerance in Jatropha curcas seedlings. Acta Physiol Plant 35, 3457-3464.
DOI URL |
| [13] |
Chen C, Yu Y, Ding XD, Liu BD, Duanmu H, Zhu D, Sun XL, Cao L, Nisa ZU, Li Q, Zhu YM (2018). Genome-wide analysis and expression profiling of PP2C clade D under saline and alkali stresses in wild soybean and Arabidopsis. Protoplasma 255, 643-654.
DOI URL |
| [14] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI URL |
| [15] |
Clark K, Franco JY, Schwizer S, Pang Z, Hawara E, Liebrand T (2018). An effector from the huanglongbing-associated pathogen targets citrus proteases. Nat Commun 9, 1718.
DOI URL |
| [16] |
Dando PM, Fortunato M, Strand GB, Smith TS, Barrett AJ (2003). Pyroglutamyl-peptidase I: cloning, sequencing, and characterisation of the recombinant human enzyme. Protein Expr Purif 28, 111-119.
DOI URL |
| [17] |
Earnshaw WC, Martins LM, Kaufmann SH (1999). Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68, 383-424.
PMID |
| [18] |
Finn RD, Penelope C, Eberhardt RY, Eddy SR, Jaina M, Mitchell AL, Potter SC, Marco P, Matloob Q, Amaia SV (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44, D279-D285.
DOI URL |
| [19] |
Grudkowska M, Zagdanska B (2004). Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51, 609-624.
DOI URL |
| [20] |
Hara-Nishimura I (1995). Vacuolar processing enzyme responsible for maturation of vacuolar proteins. Seikagaku 67, 372-377.
PMID |
| [21] |
Hatsugai N, Kuroyanagi M, Nishimura M, Hara-Nishimura I (2006). A cellular suicide strategy of plants: vacuole- mediated cell death. Apoptosis 11, 905-911.
PMID |
| [22] |
Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I (2005). Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J Biol Chem 280, 32914-32920.
PMID |
| [23] |
Li ZG, Zeng HZ, Ao PX, Gong M (2014). Lipid response to short-term chilling shock and long-term chill hardening in Jatropha curcas L. seedlings. Acta Physiol Plant 36, 2803-2814.
DOI URL |
| [24] |
Liu HJ, Hu MH, Wang Q, Cheng L, Zhang ZB (2018). Role of papain-like cysteine proteases in plant development. Front Plant Sci 9, 1717.
DOI URL |
| [25] |
Megha S, Basu U, Kav NNV (2018). Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ 41, 1-15.
DOI URL |
| [26] |
Montes JM, Melchinger AE (2016). Domestication and breeding of Jatropha curcas L. Trend Plant Sci 21, 1045-1057.
DOI URL |
| [27] |
Niño MC, Kim MS, Kang KK, Cho YG (2020). Genome- wide identification and molecular characterization of cysteine protease genes in rice. Plant Biotechnol Rep 14, 69-87.
DOI URL |
| [28] |
Rawlings ND, Barrett AJ, Bateman A (2010). MEROPS: the peptidase database. Nucl Acids Res 38, D227-D233.
DOI URL |
| [29] | Rawlings ND, Salvesen G (2013). Handbook of Proteolytic Enzymes. Salt Lake City: Academic Press. pp. 1253-1257. |
| [30] |
Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RAL (2012). Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol 158, 1583-1599.
DOI URL |
| [31] |
Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, Kobayashi M, Kondo M, Nishimura M, Hara-Nishimura I (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278, 32292-32299.
PMID |
| [32] |
Stroeher VL, Maclagan JL, Good AG (1997). Molecular cloning of a Brassica napus cysteine protease gene inducible by drought and low temperature stress. Physiol Plant 101, 389-397.
DOI URL |
| [33] |
Vorster BJ, Cullis CA, Kunert KJ (2019). Plant vacuolar processing enzymes. Front Plant Sci 10, 479.
DOI PMID |
| [34] |
Wang HB, Gong M, Guo JY, Xin H, Gao Y, Liu C, Dai DQ, Tang LZ (2018a). Genome-wide Identification of Jatropha curcas MAPK, MAPKK, and MAPKKK gene families and their expression profile under cold stress. Sci Rep 8, 16163.
DOI URL |
| [35] |
Wang HB, Zou ZR, Wang SS, Gong M (2013a). Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One 8, e82817.
DOI URL |
| [36] | Wang SS, Wang HB, Gong M (2013b). Identification of microRNAs involved in chilling response by deep sequencing of Jatropha curcas L. small RNAs at the global genome level. In: Proceedings of the 21st European Biomass Conference and Exhibition. Copenhagen: European Union. pp. 356-363. |
| [37] |
Wang W, Zhou XM, Xiong HX, Mao WY, Zhao P, Sun MX (2018b). Papain-like and legumain-like proteases in rice: genome-wide identification, comprehensive gene feature characterization and expression analysis. BMC Plant Biol 18, 87.
DOI PMID |
| [38] |
Wani SH, Kumar V, Khare T, Tripathi P, Shah T, Ramakrishna C, Aglawe S, Mangrauthia SK (2020). miRNA applications for engineering abiotic stress tolerance in plants. Biologia 75, 1063-1081.
DOI URL |
| [39] |
Wilkinson KD, Laleli-Sahin E, Urbauer J, Larsen CN, Shih GH, Haas AL, Walsh STR, Wand AJ (1999). The binding site for UCH-L3 on ubiquitin: mutagenesis and NMR studies on the complex between ubiquitin and UCH- L3. J Mol Biol 291, 1067-1077.
PMID |
| [40] |
Wu PZ, Zhou CP, Cheng SF, Wu ZY, Lu WJ, Han JL, Chen YB, Chen Y, Ni PX, Wang Y, Xu X, Huang Y, Song C, Wang ZW, Shi N, Zhang XD, Fang XH, Yang Q, Jiang HW, Chen YP, Li MR, Wang Y, Chen F, Wang J, Wu GJ (2015). Integrated genome sequence and linkage map of physic nut ( Jatropha curcas L), a biodiesel plant. Plant J 81, 810-821.
DOI URL |
| [41] |
Zang QW, Wang CX, Li XY, Guo ZA, Jing RL, Zhao J, Chang XP (2010). Isolation and characterization of a gene encoding a polyethylene glycol-induced cysteine protease in common wheat. J Biosci 35, 379-388.
DOI URL |
| [42] |
Zhang X, Pan BZ, Chen MS, Chen W, Li J, Xu ZF, Liu CN (2019). JCDB: a comprehensive knowledge base for Jatropha curcas, an emerging model for woody energy plants. BMC Genomics 20, 958.
DOI PMID |
| [43] |
Zheng L, Chen SS, Xie LH, Lu ZC, Liu MY, Han XJ, Qiao GR, Jiang J, Zhuo RY, Qiu WM, He ZQ (2018). Overexpression of cysteine protease gene from Salix matsudana enhances salt tolerance in transgenic Arabidopsis. Environ Exp Bot 147, 53-62.
DOI URL |
| [44] |
Zou Z, Huang QX, Xie GS, Yang LF (2018). Genome-wide comparative analysis of papain-like cysteine protease family genes in castor bean and physic nut. Sci Rep 8, 331.
DOI URL |
| [1] | Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli. Identification of the Spinach AT-hook Gene Family and Analysis of Expression Profiles [J]. Chinese Bulletin of Botany, 2025, 60(3): 377-392. |
| [2] | Jiaqi Gu, Fuhui Zhu, Peihao Xie, Qingying Meng, Ying Zheng, Xianlong Zhang, Daojun Yuan. Genome-wide Identification and Domestication Analysis of the Phytochrome PHY Gene Family in Gossypium [J]. Chinese Bulletin of Botany, 2024, 59(1): 34-53. |
| [3] | Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang. Phylogenetic and Expression Analysis of MADS-box Gene Family in Rhododendron ovatum [J]. Chinese Bulletin of Botany, 2023, 58(3): 404-416. |
| [4] | Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149. |
| [5] | Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang. Evolution and Expression of NCED Family Genes in Vitis vinifera [J]. Chinese Bulletin of Botany, 2019, 54(4): 474-485. |
| [6] | Kang Tang,Ruolin Yang. Origin and Evolution of Soybean Protein-coding Genes [J]. Chinese Bulletin of Botany, 2019, 54(3): 316-327. |
| [7] | Zheng Jun, Qiao Ling, Zhao Jiajia, Qiao Linyi, Zhang Shichang, Chang Jianzhong, Tang Caiguo, Yang Sanwei. Whole-genome Analysis of CCT Gene Family and Their Responses to Phytohormones in Aegilops tauschii [J]. Chinese Bulletin of Botany, 2017, 52(2): 188-201. |
| [8] | Fangzheng Li, Suxin Yang, Chunxia Wu, Haichao Wei, Ruilian Qu, Xianzhong Feng. Structure and Expression Analysis of KNOX Gene Family in Soybean [J]. Chinese Bulletin of Botany, 2012, 47(3): 236-247. |
| [9] | Ying Li;Kaijing Zuo;Kexuan Tang. A Survey of Functional Studies of the GH3 Gene Family in Plants [J]. Chinese Bulletin of Botany, 2008, 25(05): 507-515. |
| [10] | Jian Zhang;Guixia Xu;Haoyue Xue;Jin Hu. Foundation and Current Progress of Plant Evolutionary Developmental Biology [J]. Chinese Bulletin of Botany, 2007, 24(01): 1-30. |
| [11] | GUO Shang-Jing CHEN Na MENG Qing-Wei. Chloroplast-localized Small Heat Shock Proteins [J]. Chinese Bulletin of Botany, 2005, 22(02): 223-230. |
| [12] | LIU Zhi-Min, CHEN Jun-Jie, YANG Shao-Hua, . Molecular diversity of glutamate receptors and its molecular mechanism [J]. Biodiv Sci, 1999, 07(1): 52-59. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||