

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (3): 316-327.DOI: 10.11983/CBB18176 cstr: 32102.14.CBB18176
• RESEARCH ARTICLE • Previous Articles Next Articles
Received:2018-08-14
Accepted:2018-12-10
Online:2019-05-01
Published:2019-11-24
Contact:
Ruolin Yang
Kang Tang,Ruolin Yang. Origin and Evolution of Soybean Protein-coding Genes[J]. Chinese Bulletin of Botany, 2019, 54(3): 316-327.
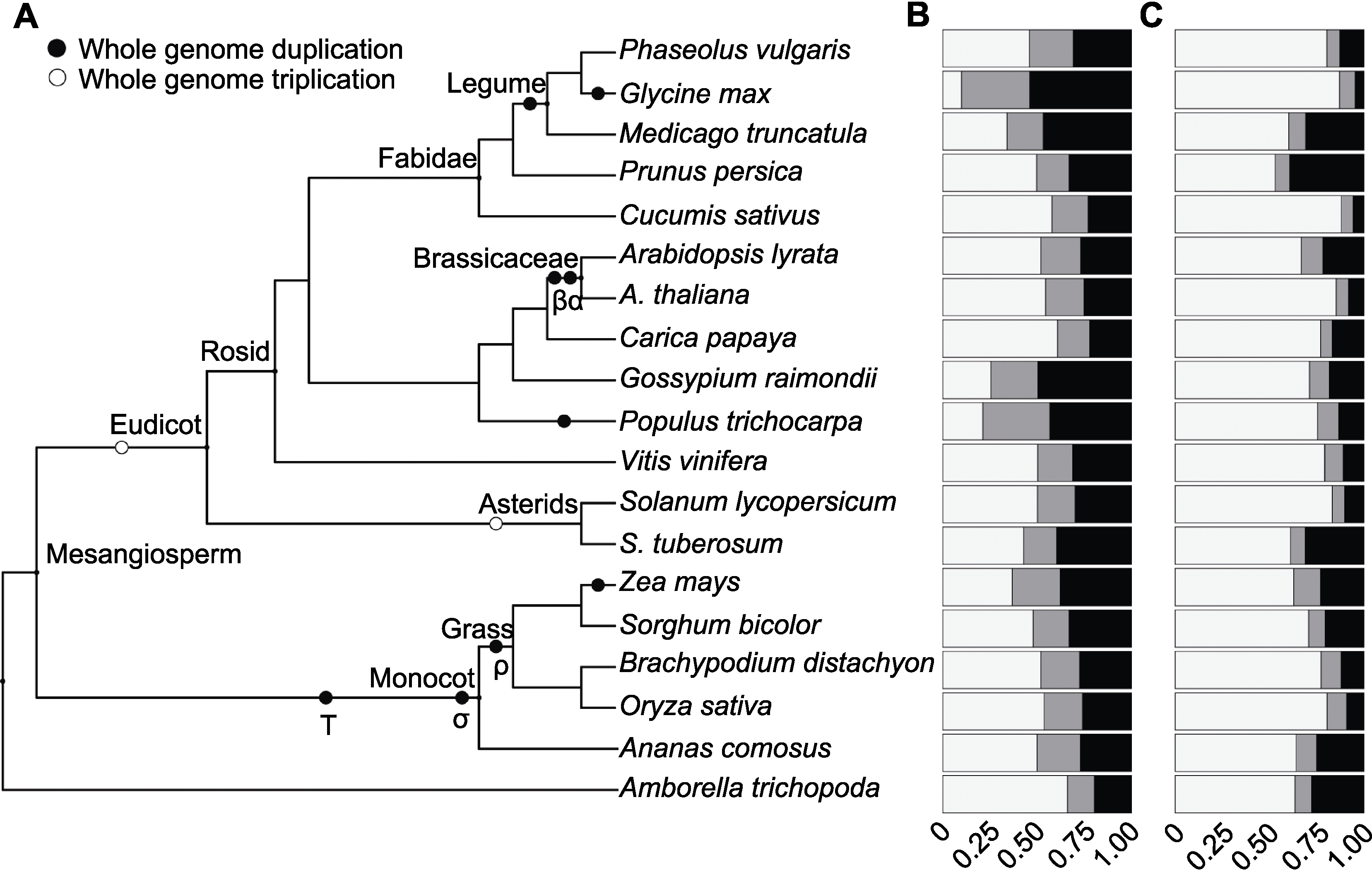
Figure 1 Gene family size distribution of 19 angiosperm species(A) Phylogenetic tree showing the relationships between the 19 angiosperm species used in this study; (B) Homologous gene family sizes; (C) Gene family sizes of orphan genes. The colors indicate the proportions of genes, white for singletons, grey for two-genes and black for multigenes.
| Species | Singletons | Two-gene families | Multigene families | Total gene families | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 9823 | 1061(2122) | 523(2935) | 11407 | 207 |
| Ananas comosus | 9059 | 2087(4174) | 1007(4916) | 12153 | 124 |
| Oryza sativa | 11966 | 2269(4538) | 1167(5805) | 15402 | 64 |
| Brachypodium distachyon | 11455 | 2264(4528) | 1209(6066) | 14928 | 50 |
| Sorghum bicolor | 12663 | 2529(5058) | 1399(8749) | 16591 | 416 |
| Zea mays | 10277 | 3568(7136) | 1964(10539) | 15809 | 297 |
| Solanum tuberosum | 11592 | 2390(4780) | 1399(10741) | 15381 | 1051 |
| S. lycopersicum | 12210 | 2448(4896) | 1371(7277) | 16029 | 72 |
| Vitis vinifera | 10408 | 1931(3862) | 1104(6483) | 13443 | 100 |
| Populus trichocarpa | 6550 | 5476(10952) | 2337(13368) | 14363 | 108 |
| Gossypium raimondii | 7582 | 3700(7400) | 2960(14806) | 14242 | 90 |
| Carica papaya | 10776 | 1505(3010) | 667(3948) | 12948 | 194 |
| Arabidopsis thaliana | 13278 | 2485(4970) | 1194(6144) | 16957 | 125 |
| A. lyrata | 12767 | 2605(5210) | 1327(6596) | 16699 | 67 |
| Cucumis sativus | 10152 | 1691(3382) | 795(4038) | 12638 | 38 |
| Prunus persica | 10822 | 1876(3752) | 1106(7192) | 13804 | 217 |
| Medicago truncatula | 9936 | 2812(5624) | 1948(13673) | 14696 | 308 |
| Glycine max | 4241 | 7735(15470) | 4206(23027) | 16182 | 153 |
| Phaseolus vulgaris | 11324 | 2873(5746) | 1430(7626) | 15569 | 132 |
Table 1 Number of homologous gene families (and genes) in 19 angiosperm species
| Species | Singletons | Two-gene families | Multigene families | Total gene families | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 9823 | 1061(2122) | 523(2935) | 11407 | 207 |
| Ananas comosus | 9059 | 2087(4174) | 1007(4916) | 12153 | 124 |
| Oryza sativa | 11966 | 2269(4538) | 1167(5805) | 15402 | 64 |
| Brachypodium distachyon | 11455 | 2264(4528) | 1209(6066) | 14928 | 50 |
| Sorghum bicolor | 12663 | 2529(5058) | 1399(8749) | 16591 | 416 |
| Zea mays | 10277 | 3568(7136) | 1964(10539) | 15809 | 297 |
| Solanum tuberosum | 11592 | 2390(4780) | 1399(10741) | 15381 | 1051 |
| S. lycopersicum | 12210 | 2448(4896) | 1371(7277) | 16029 | 72 |
| Vitis vinifera | 10408 | 1931(3862) | 1104(6483) | 13443 | 100 |
| Populus trichocarpa | 6550 | 5476(10952) | 2337(13368) | 14363 | 108 |
| Gossypium raimondii | 7582 | 3700(7400) | 2960(14806) | 14242 | 90 |
| Carica papaya | 10776 | 1505(3010) | 667(3948) | 12948 | 194 |
| Arabidopsis thaliana | 13278 | 2485(4970) | 1194(6144) | 16957 | 125 |
| A. lyrata | 12767 | 2605(5210) | 1327(6596) | 16699 | 67 |
| Cucumis sativus | 10152 | 1691(3382) | 795(4038) | 12638 | 38 |
| Prunus persica | 10822 | 1876(3752) | 1106(7192) | 13804 | 217 |
| Medicago truncatula | 9936 | 2812(5624) | 1948(13673) | 14696 | 308 |
| Glycine max | 4241 | 7735(15470) | 4206(23027) | 16182 | 153 |
| Phaseolus vulgaris | 11324 | 2873(5746) | 1430(7626) | 15569 | 132 |
| Species | Singletons | Two-gene families | Multigene families | Species-specific genes | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 7892 | 547(1094) | 502(3447) | 12433 | 105 |
| Ananas comosus | 5685 | 483(966) | 297(2224) | 8875 | 94 |
| Oryza sativa | 10774 | 686(1372) | 292(1224) | 13370 | 29 |
| Brachypodium distachyon | 3485 | 235(470) | 125(548) | 4503 | 15 |
| Sorghum bicolor | 5682 | 350(700) | 254(1644) | 8026 | 103 |
| Zea mays | 7253 | 813(1626) | 552(2643) | 11522 | 65 |
| Solanum tuberosum | 7278 | 471(942) | 376(3688) | 11908 | 163 |
| S. lycopersicum | 7836 | 308(616) | 177(950) | 9402 | 51 |
| Vitis vinifera | 7238 | 445(890) | 229(1006) | 9134 | 44 |
| Populus trichocarpa | 7923 | 593(1186) | 281(1398) | 10507 | 31 |
| Gossypium raimondii | 5495 | 408(816) | 293(1406) | 7717 | 26 |
| Carica papaya | 7680 | 307(614) | 224(1653) | 9947 | 88 |
| Arabidopsis thaliana | 2751 | 105(210) | 57(261) | 3222 | 21 |
| A. lyrata | 5413 | 461(922) | 366(1759) | 8094 | 83 |
| Cucumis sativus | 3458 | 125(250) | 54(223) | 3931 | 13 |
| Prunus persica | 3347 | 242(484) | 195(2483) | 6314 | 838 |
| Medicago truncatula | 12763 | 962(1924) | 820(6524) | 21211 | 145 |
| Glycine max | 9961 | 476(952) | 118(523) | 11436 | 23 |
| Phaseolus vulgaris | 2013 | 85(170) | 58(318) | 2501 | 19 |
Table 2 Number of orphan gene families (and genes) in 19 angiosperm species
| Species | Singletons | Two-gene families | Multigene families | Species-specific genes | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 7892 | 547(1094) | 502(3447) | 12433 | 105 |
| Ananas comosus | 5685 | 483(966) | 297(2224) | 8875 | 94 |
| Oryza sativa | 10774 | 686(1372) | 292(1224) | 13370 | 29 |
| Brachypodium distachyon | 3485 | 235(470) | 125(548) | 4503 | 15 |
| Sorghum bicolor | 5682 | 350(700) | 254(1644) | 8026 | 103 |
| Zea mays | 7253 | 813(1626) | 552(2643) | 11522 | 65 |
| Solanum tuberosum | 7278 | 471(942) | 376(3688) | 11908 | 163 |
| S. lycopersicum | 7836 | 308(616) | 177(950) | 9402 | 51 |
| Vitis vinifera | 7238 | 445(890) | 229(1006) | 9134 | 44 |
| Populus trichocarpa | 7923 | 593(1186) | 281(1398) | 10507 | 31 |
| Gossypium raimondii | 5495 | 408(816) | 293(1406) | 7717 | 26 |
| Carica papaya | 7680 | 307(614) | 224(1653) | 9947 | 88 |
| Arabidopsis thaliana | 2751 | 105(210) | 57(261) | 3222 | 21 |
| A. lyrata | 5413 | 461(922) | 366(1759) | 8094 | 83 |
| Cucumis sativus | 3458 | 125(250) | 54(223) | 3931 | 13 |
| Prunus persica | 3347 | 242(484) | 195(2483) | 6314 | 838 |
| Medicago truncatula | 12763 | 962(1924) | 820(6524) | 21211 | 145 |
| Glycine max | 9961 | 476(952) | 118(523) | 11436 | 23 |
| Phaseolus vulgaris | 2013 | 85(170) | 58(318) | 2501 | 19 |
| Phylostratum internode | Genes (%) | Singletons | Two-genes | Multigenes |
|---|---|---|---|---|
| Angiosperm (PS1) | 30932(58.7%) | 1982 | 5150(10300) | 3400(18650) |
| Mesangiosperm (PS2) | 4057(7.7%) | 508 | 708(1416) | 359(2133) |
| Eudicot (PS3) | 2356(4.5%) | 303 | 521(1042) | 206(1011) |
| Rosid (PS4) | 582(1.1%) | 109 | 181(362) | 31(111) |
| Legume (PS5) | 1780(3.4%) | 460 | 452(904) | 87(416) |
| Phaseoleae (PS6) | 1590(3.0%) | 568 | 400(800) | 49(222) |
| Soybean (PS7) | 11436(21.7%) | 9961 | 476(952) | 118(523) |
Table 3 Number of soybean gene families (and genes) assigned to each phylostratum
| Phylostratum internode | Genes (%) | Singletons | Two-genes | Multigenes |
|---|---|---|---|---|
| Angiosperm (PS1) | 30932(58.7%) | 1982 | 5150(10300) | 3400(18650) |
| Mesangiosperm (PS2) | 4057(7.7%) | 508 | 708(1416) | 359(2133) |
| Eudicot (PS3) | 2356(4.5%) | 303 | 521(1042) | 206(1011) |
| Rosid (PS4) | 582(1.1%) | 109 | 181(362) | 31(111) |
| Legume (PS5) | 1780(3.4%) | 460 | 452(904) | 87(416) |
| Phaseoleae (PS6) | 1590(3.0%) | 568 | 400(800) | 49(222) |
| Soybean (PS7) | 11436(21.7%) | 9961 | 476(952) | 118(523) |
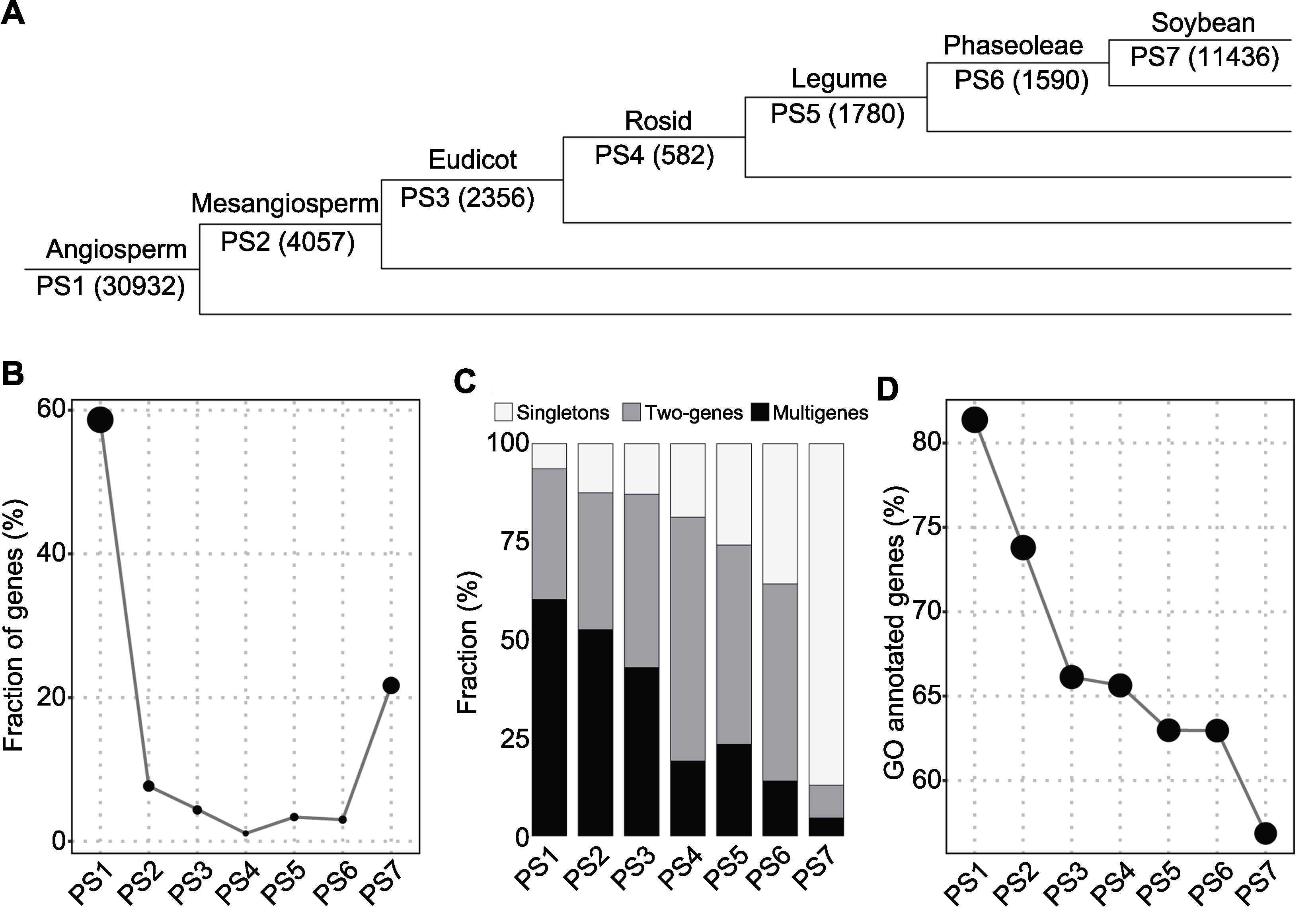
Figure 2 Origination of soybean genes(A) Numbers in parenthesis denote the number of genes per phylostratum (PS1-PS7); (B) Gene fraction; (C) Gene copy status; (D) Gene Ontology annotation
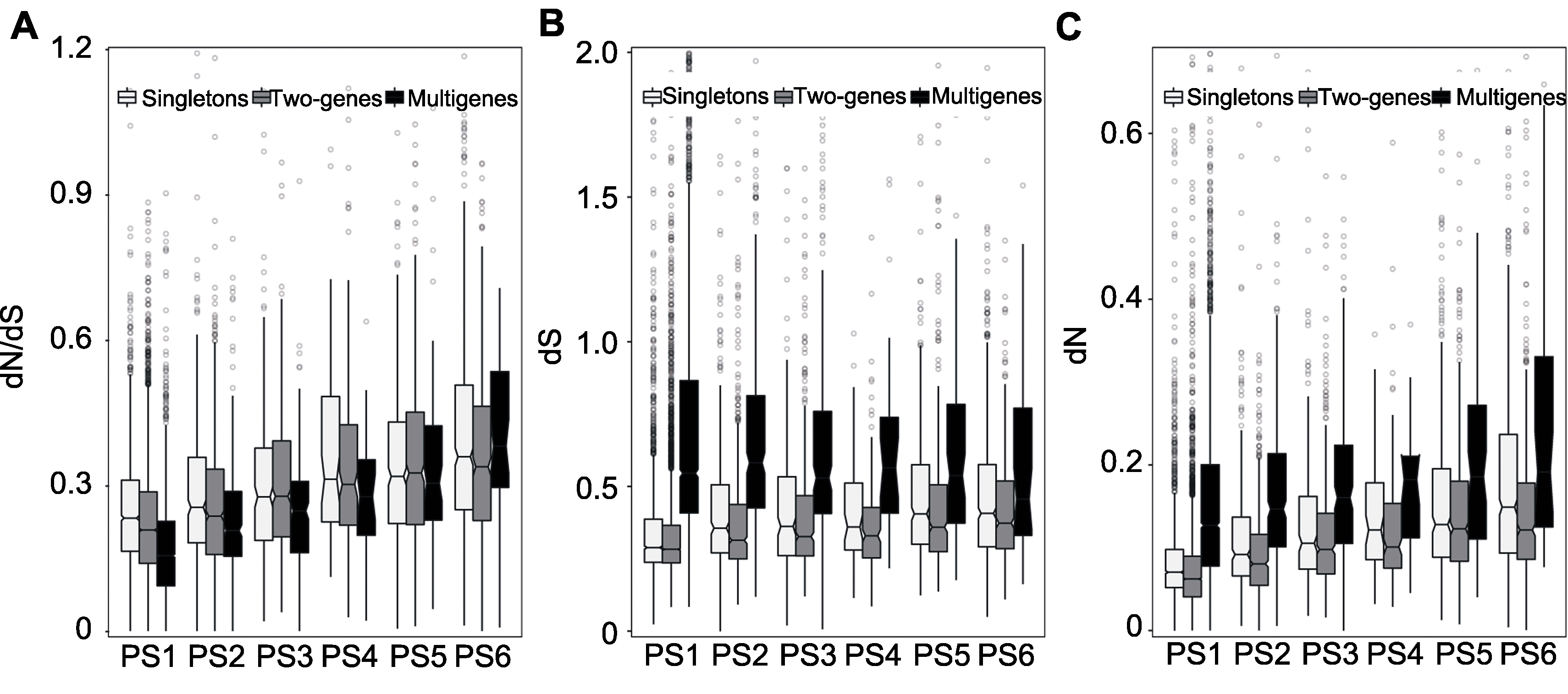
Figure 3 Divergence degrees of soybean genes Estimated between soybean and common bean selection pressure (dN/dS)(A), synonymous substitution rate (dS) (B) and nonsynonymous substitution rate (dN) (C).
| [1] |
孙红正, 葛颂 ( 2010). 重复基因的进化——回顾与进展. 植物学报 45, 13-22.
DOI URL |
| [2] | Albalat R, Cañestro C ( 2016). Evolution by gene loss. Nat Rev Genet 17, 379-391. |
| [3] |
Amborella Genome Project ( 2013). The Amborella genome and the evolution of flowering plants. Science 342, 124-1089.
DOI URL PMID |
| [4] |
Bolger AM, Lohse M, Usadel B ( 2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
DOI URL PMID |
| [5] |
Cai JJ, Borenstein E, Chen R, Petrov DA ( 2009). Similarly strong purifying selection acts on human disease genes of all evolutionary ages. Genome Biol Evol 1, 131-144.
DOI URL PMID |
| [6] |
Chen SD, Krinsky BH, Long MY ( 2013). New genes as drivers of phenotypic evolution. Nat Rev Genet 14, 645-660.
DOI URL PMID |
| [7] |
Chen TW, Wu TH, Ng WV, Lin WC ( 2011). Interrogation of alternative splicing events in duplicated genes during evolution. BMC Genomics 12(Suppl3), S16.
DOI URL PMID |
| [8] |
Domazet-Lošo T, Brajković J, Tautz D ( 2007). A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23, 533-539.
DOI URL PMID |
| [9] |
Doyle JJ, Luckow MA ( 2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131, 900-910.
DOI URL |
| [10] |
Enright AJ, Van Dongen S, Ouzounis CA ( 2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575-1584.
URL PMID |
| [11] |
Foissac S, Sammeth M ( 2007). ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35, W297-W299.
DOI URL PMID |
| [12] |
Freeling M ( 2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60, 433-453.
DOI URL |
| [13] |
Guo YL ( 2013). Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J 73, 941-951.
DOI URL PMID |
| [14] |
Jiao YN, Paterson AH ( 2014). Polyploidy-associated genome modifications during land plant evolution. Philos Trans R Soc Lond B Biol Sci 369, 20130355.
DOI URL PMID |
| [15] |
Kaessmann H ( 2010). Origins, evolution, and phenotypic impact of new genes. Genome Res 20, 1313-1326.
DOI URL |
| [16] |
Keren H, Lev-Maor G, Ast G ( 2010). Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11, 345-355.
DOI URL PMID |
| [17] |
Kim D, Langmead B, Salzberg SL ( 2015). HISAT: a fast spliced aligner with low memory requirements. Nat Me- thods 12, 357-360.
DOI URL PMID |
| [18] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG ( 2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948.
DOI URL |
| [19] |
Li L, Stoeckert CJ Jr, Roos DS ( 2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13, 2178-2189.
DOI URL |
| [20] | Long M, Betrán E, Thornton K, Wang W ( 2003). The origin of new genes: glimpses from the young and old. Nat Rev Genet 4, 865-875. |
| [21] |
Lynch M, Conery JS ( 2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151-1155.
DOI URL PMID |
| [22] |
Merkin J, Russell C, Chen P, Burge CB ( 2012). Evolutionary dynamics of gene and isoform regulation in mam- malian tissues. Science 338, 1593-1599.
DOI URL PMID |
| [23] |
Michael TP, Jackson S ( 2013). The first 50 plant genomes. Plant Gen 6, 2.
DOI URL |
| [24] |
Michael TP, VanBuren R ( 2015). Progress, challenges and the future of crop genomes. Curr Opin Plant Biol 24, 71-81.
DOI URL PMID |
| [25] | Ohno S ( 1970). Evolution by Gene Duplication. Berlin, Heidelberg: Springer. pp. 1-160. |
| [26] |
Panchy N, Lehti-Shiu M, Shiu SH ( 2016). Evolution of gene duplication in plants. Plant Physiol 171, 2294-2316.
DOI URL PMID |
| [27] |
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL ( 2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290-295.
DOI URL PMID |
| [28] |
Quint M, Drost HG, Gabel A, Ullrich KK, Bönn M, Grosse I ( 2012). A transcriptomic hourglass in plant embryogenesis. Nature 490, 98-101.
DOI URL PMID |
| [29] |
Reddy ASN, Marquez Y, Kalyna M, Barta A ( 2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657-3683.
DOI URL |
| [30] |
Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA ( 2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178-183.
DOI URL |
| [31] |
Shen YT, Zhou ZK, Wang Z, Li WY, Fang C, Wu M, Ma YM, Liu TF, Kong LA, Peng DL, Tian ZX ( 2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996-1008.
DOI URL PMID |
| [32] |
Suyama M, Torrents D, Bork P ( 2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609-W612.
DOI URL PMID |
| [33] |
Tasdighian S, Van Bel M, Li Z, Van de Peer Y, Carretero-Paulet L, Maere S ( 2017). Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell 29, 2766-2785.
DOI URL PMID |
| [34] |
Tautz D, Domazet-Lošo T ( 2011). The evolutionary origin of orphan genes. Nat Rev Genet 12, 692-702.
DOI URL PMID |
| [35] |
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L ( 2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511-515.
DOI URL PMID |
| [36] |
Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O ( 2005). Genome-wide midrange transcription profiles reveal expression level re- lationships in human tissue specification. Bioinformatics 21, 650-659.
DOI URL PMID |
| [37] |
Yang ZH ( 2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586-1591.
DOI URL PMID |
| [38] |
Zhang JZ ( 2003). Evolution by gene duplication: an update. Trends Ecol Evol 18, 292-298.
DOI URL |
| [1] | WANG Xiu-Yuan, SHEN Lei, LIU Ting-Ting, WEI Wen-Wen, ZHANG Shuai, ZHANG Wei. Spatial and temporal distribution of root system and interspecific competition strategy in Malus pumila ‘Saiwaihong’ - Glycine max agroforestry system [J]. Chin J Plant Ecol, 2025, 49(5): 748-759. |
| [2] | Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli. Identification of the Spinach AT-hook Gene Family and Analysis of Expression Profiles [J]. Chinese Bulletin of Botany, 2025, 60(3): 377-392. |
| [3] | Jie Cao, Qiulian Lu, Jianping Zhai, Baohui Liu, Chao Fang, Shichen Li, Tong Su. Changes in the Expression of the Soybean TPS Gene Family Under Salt Stress and Haplotype Selection Analysis [J]. Chinese Bulletin of Botany, 2025, 60(2): 172-185. |
| [4] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [5] | Jiaxin Chen, Hao Mei, Caixiang Huang, Zongyuan Liang, Yitong Quan, Dongpeng Li, Buweimaieryemu·Saimaiti , Xinxin Li, Hong Liao. A Highly Efficient Method to Generate Chimeric Soybean Plant with Transgenic Hairy Roots [J]. Chinese Bulletin of Botany, 2024, 59(1): 89-98. |
| [6] | Feifei Zhang, Tianfeng Yang, Lirong Chen, Dongmei Liu, Liuyuan Yang, Duyu Yang, Peng Ju, Lu Lu. Review of pollen color diversity in Angiosperms [J]. Biodiv Sci, 2024, 32(1): 23346-. |
| [7] | Jiaqi Gu, Fuhui Zhu, Peihao Xie, Qingying Meng, Ying Zheng, Xianlong Zhang, Daojun Yuan. Genome-wide Identification and Domestication Analysis of the Phytochrome PHY Gene Family in Gossypium [J]. Chinese Bulletin of Botany, 2024, 59(1): 34-53. |
| [8] | Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang. Phylogenetic and Expression Analysis of MADS-box Gene Family in Rhododendron ovatum [J]. Chinese Bulletin of Botany, 2023, 58(3): 404-416. |
| [9] | Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149. |
| [10] | Shumei Zhang, Wei Li, Dingnan Li. Inventory of species diversity of Liaoning higher plants [J]. Biodiv Sci, 2022, 30(6): 22038-. |
| [11] | Yunhui Wang, Yifan Wang, Jiayu Lin, Jinhong Li, Shien Yao, Xiangchi Feng, Zhenlin Cao, Jun Wang, Meina Li. Plant Kinesin: from Microtubule Arrays to Physiological Regulation [J]. Chinese Bulletin of Botany, 2022, 57(3): 358-374. |
| [12] | Dandan Wu, Yongkun Chen, Yu Yang, Chunyan Kong, Ming Gong. Identification of the Cysteine Protease Family and Corresponding miRNAs in Jatropha curcas and Their Response to Chill-hardening [J]. Chinese Bulletin of Botany, 2021, 56(5): 544-558. |
| [13] | Mengke Du, Wenting Lian, Xiao Zhang, Xinxin Li. Effects of Nitrogen Application on Nitrogen Fixation Capacity and GmLbs Expression in Soybean [J]. Chinese Bulletin of Botany, 2021, 56(4): 391-403. |
| [14] | Zhengjun Xia, Yuzhuo Li, Jinlong Zhu, Hongyan Wu, Kun Xu, Hong Zhai. A Rapid, Non-destructive and Continuous Sampling Technique and DNA Extraction for Soybean Seed [J]. Chinese Bulletin of Botany, 2021, 56(1): 56-61. |
| [15] | Yan Wang, Bowei Jia, Mingzhe Sun, Xiaoli Sun. Advances in Molecular Mechanisms of Stress Tolerance in Wild Soybean [J]. Chinese Bulletin of Botany, 2021, 56(1): 104-115. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
