

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (3): 296-299.DOI: 10.11983/CBB19033 cstr: 32102.14.CBB19033
• COMMENTARIES • Previous Articles Next Articles
Received:2019-01-21
Accepted:2019-01-22
Online:2019-05-01
Published:2019-05-20
Contact:
Kabin Xie
Kabin Xie. Chinese Scientists Reveal Genome-wide Off-targeted Editing of Cytosine Base Editor[J]. Chinese Bulletin of Botany, 2019, 54(3): 296-299.
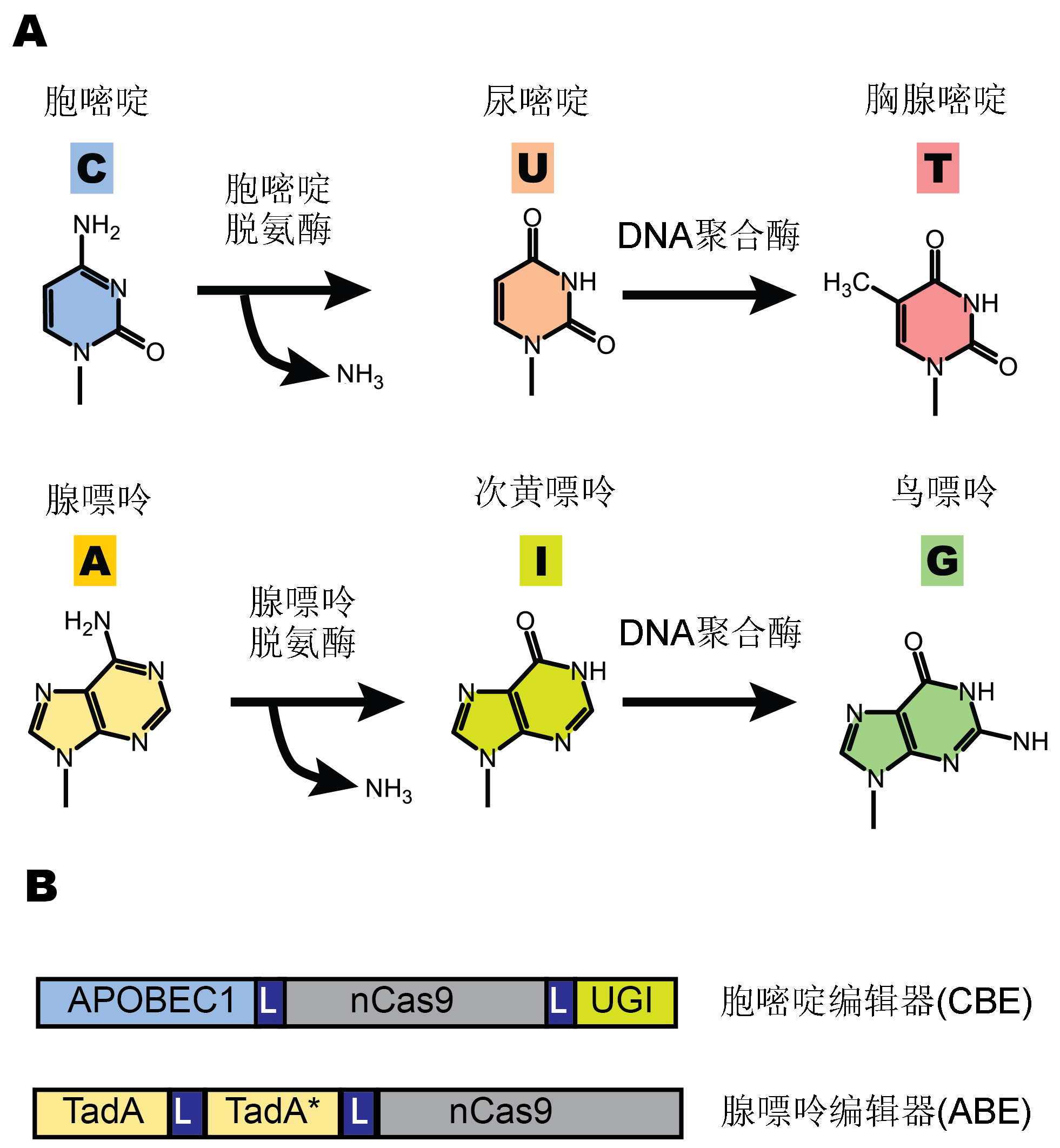
Figure 1 CRISRP-Cas9 base editing(A) Deaminase-mediated base editing reactions; (B) The schematic structure of cytosine (CBE, take BE3 as an example) and adenine (ABE) base editors. L: Peptide linker
| [1] |
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR ( 2017). Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551, 464-471.
DOI URL PMID |
| [2] |
Hua K, Tao X, Yuan F, Wang D, Zhu JK ( 2018). Precise A·T to G·C base editing in the rice genome. Mol Plant 11, 627-630.
DOI URL |
| [3] | Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu J, Zhang F, Gao C ( 2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292-295. |
| [4] |
Kang BC, Yun JY, Kim ST, Shin Y, Ryu J, Choi M, Woo JW, Kim JS ( 2018). Precision genome engineering th- rough adenine base editing in plants. Nat Plants 4, 427-431.
DOI URL |
| [5] |
Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR ( 2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371-376.
DOI URL PMID |
| [6] |
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR ( 2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420-424.
DOI URL PMID |
| [7] |
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C ( 2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19, 59.
DOI URL |
| [8] |
Li J, Sun Y, Du J, Zhao Y, Xia L ( 2017). Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10, 526-529.
DOI URL PMID |
| [9] |
Lu Y, Zhu JK ( 2017). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant 10, 523-525.
DOI URL PMID |
| [10] |
Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X ( 2016). Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Me- thods 13, 1029-1035.
DOI URL PMID |
| [11] |
May A ( 2017). Base editing on the rise. Nat Biotechnol 35, 428-429.
DOI URL PMID |
| [12] |
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A ( 2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729.
DOI URL PMID |
| [13] |
Rees HA, Liu DR ( 2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770-788.
DOI |
| [14] |
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H ( 2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant 11, 623-626.
DOI URL PMID |
| [15] |
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A ( 2017). Targeted base editing in rice and tomato using a CRISPR-1)Cas9 cytidine deaminase fusion. Nat Biotechnol 35, 441-443.
DOI URL PMID |
| [16] |
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H ( 2018). Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11, 631-634.
DOI URL |
| [17] |
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C ( 2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438-440.
DOI URL PMID |
| [18] | Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H ( 2019). Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289-292. |
| [1] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [2] | Danling Hu, Yongwei Sun. Advances in Virus-mediated Genome Editing Technology in Plants [J]. Chinese Bulletin of Botany, 2024, 59(3): 452-462. |
| [3] | Chunyan Miao, Mingming Li, Xin Zuo, Ning Ding, Jiafang Du, Juan Li, Zhongyi Zhang, Fengqing Wang. Establishment of CRISPR/Cas9 Gene Editing System in Rehmannia henryi [J]. Chinese Bulletin of Botany, 2023, 58(6): 905-916. |
| [4] | Wenqi Zhou, Yuqian Zhou, Yongsheng Li, Haijun He, Yanzhong Yang, Xiaojuan Wang, Xiaorong Lian, Zhongxiang Liu, Zhubing Hu. ZmICE2 Regulates Stomatal Development in Maize [J]. Chinese Bulletin of Botany, 2023, 58(6): 866-881. |
| [5] | He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng. Advance in Gene-editing Technology Based on CRISPR/Cas9 and Its Application in Plants [J]. Chinese Bulletin of Botany, 2022, 57(4): 508-531. |
| [6] | Yulong He, Jiage Wang, Shanshan Zhao, Jin Gao, Yingying Chang, Xiting Zhao, Bihua Nie, Qingxiang Yang, Jiangli Zhang, Mingjun Li. Establishment and Application of RPA-CRISPR/Cas12a Detection System for Potato Virus Y [J]. Chinese Bulletin of Botany, 2022, 57(3): 308-319. |
| [7] | Rui Zhang, Caixia Gao. Saturation Mutagenesis Using Dual Cytosine and Adenine Base Editors [J]. Chinese Bulletin of Botany, 2021, 56(1): 50-55. |
| [8] | Xianrong Xie, Dongchang Zeng, Jiantao Tan, Qinlong Zhu, Yaoguang Liu. CRISPR-based DNA Fragment Deletion in Plants [J]. Chinese Bulletin of Botany, 2021, 56(1): 44-49. |
| [9] | Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang. Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing [J]. Chinese Bulletin of Botany, 2019, 54(3): 385-395. |
| [10] | Wang Ying, Li Xianggan, Qiu Lijuan. Research Progress in Off-target in CRISPR/Cas9 Genome Editing [J]. Chinese Bulletin of Botany, 2018, 53(4): 528-541. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
