

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (6): 905-916.DOI: 10.11983/CBB22250 cstr: 32102.14.CBB22250
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Chunyan Miao1,†, Mingming Li1,†, Xin Zuo1, Ning Ding1, Jiafang Du1, Juan Li1, Zhongyi Zhang2, Fengqing Wang1,*( )
)
Received:2022-10-21
Accepted:2023-03-17
Online:2023-11-01
Published:2023-11-27
Contact:
* E-mail: fqwang@henau.edu.cn
About author:† These authors contributed equally to this paper
Chunyan Miao, Mingming Li, Xin Zuo, Ning Ding, Jiafang Du, Juan Li, Zhongyi Zhang, Fengqing Wang. Establishment of CRISPR/Cas9 Gene Editing System in Rehmannia henryi[J]. Chinese Bulletin of Botany, 2023, 58(6): 905-916.
| Primer name | Primer sequence (5′-3′) | Aim |
|---|---|---|
| RhPDS1_orf_F | CCAAAAGTTGCTCCACCTGT | Amplification of RhPDS1 from cDNA |
| RhPDS1_orf_R | TGCTGCTTCCCTATCTTCTG | |
| RhPDS1_F | ATATCAAAATGGCCCAATTCGGA | Amplification of RhPDS1 from genomic DNA |
| RhPDS1_R | TCCACACTCGAAACATATTAGGC | |
| RhPDS1q_F | GCTAGCAGATGTGGCAGC | qRT-PCR analysis of RhPDS1 |
| RhPDS1q_R | CAATGACCACTTGCAATGGC | |
| RhActin_F | GTGAGGAGATGCAGAAGCAA | qRT-PCR analysis of RhActin |
| RhActin_R | CCTCGTGTGTTCACTGACAA | |
| Rh18S_F | AGCAAGGGCAGTGTTACAAG | qRT-PCR analysis of Rh18S |
| Rh18S_R | GACCACAAGTGAGGACAAGG | |
| sgRhPDS1_F | attgAAGCAAGGGATGTGCTGGG | Construction of target site |
| sgRhPDS1_R | aaacCCCAGCACATCCCTTGCTT | |
| Detect1_F | TCAACGGCATTCGGGACAAG | Identification of the transformation |
| Detect1_R | CCACATACATATCGCGGCCA | |
| Detect2_F | TGTCCCAGGATTAGAATGATTAGGC | Identification of the transformation |
| Detect2_R | CCCAGCACATCCCTTGCTT | |
| RhPDS1t_F | CGACTATCCGAGACCAGAGC | Amplification of target region |
| RhPDS1t_R | ATGTGCAACCCAGTCTCGTA |
Table 1 Primer names and sequences
| Primer name | Primer sequence (5′-3′) | Aim |
|---|---|---|
| RhPDS1_orf_F | CCAAAAGTTGCTCCACCTGT | Amplification of RhPDS1 from cDNA |
| RhPDS1_orf_R | TGCTGCTTCCCTATCTTCTG | |
| RhPDS1_F | ATATCAAAATGGCCCAATTCGGA | Amplification of RhPDS1 from genomic DNA |
| RhPDS1_R | TCCACACTCGAAACATATTAGGC | |
| RhPDS1q_F | GCTAGCAGATGTGGCAGC | qRT-PCR analysis of RhPDS1 |
| RhPDS1q_R | CAATGACCACTTGCAATGGC | |
| RhActin_F | GTGAGGAGATGCAGAAGCAA | qRT-PCR analysis of RhActin |
| RhActin_R | CCTCGTGTGTTCACTGACAA | |
| Rh18S_F | AGCAAGGGCAGTGTTACAAG | qRT-PCR analysis of Rh18S |
| Rh18S_R | GACCACAAGTGAGGACAAGG | |
| sgRhPDS1_F | attgAAGCAAGGGATGTGCTGGG | Construction of target site |
| sgRhPDS1_R | aaacCCCAGCACATCCCTTGCTT | |
| Detect1_F | TCAACGGCATTCGGGACAAG | Identification of the transformation |
| Detect1_R | CCACATACATATCGCGGCCA | |
| Detect2_F | TGTCCCAGGATTAGAATGATTAGGC | Identification of the transformation |
| Detect2_R | CCCAGCACATCCCTTGCTT | |
| RhPDS1t_F | CGACTATCCGAGACCAGAGC | Amplification of target region |
| RhPDS1t_R | ATGTGCAACCCAGTCTCGTA |
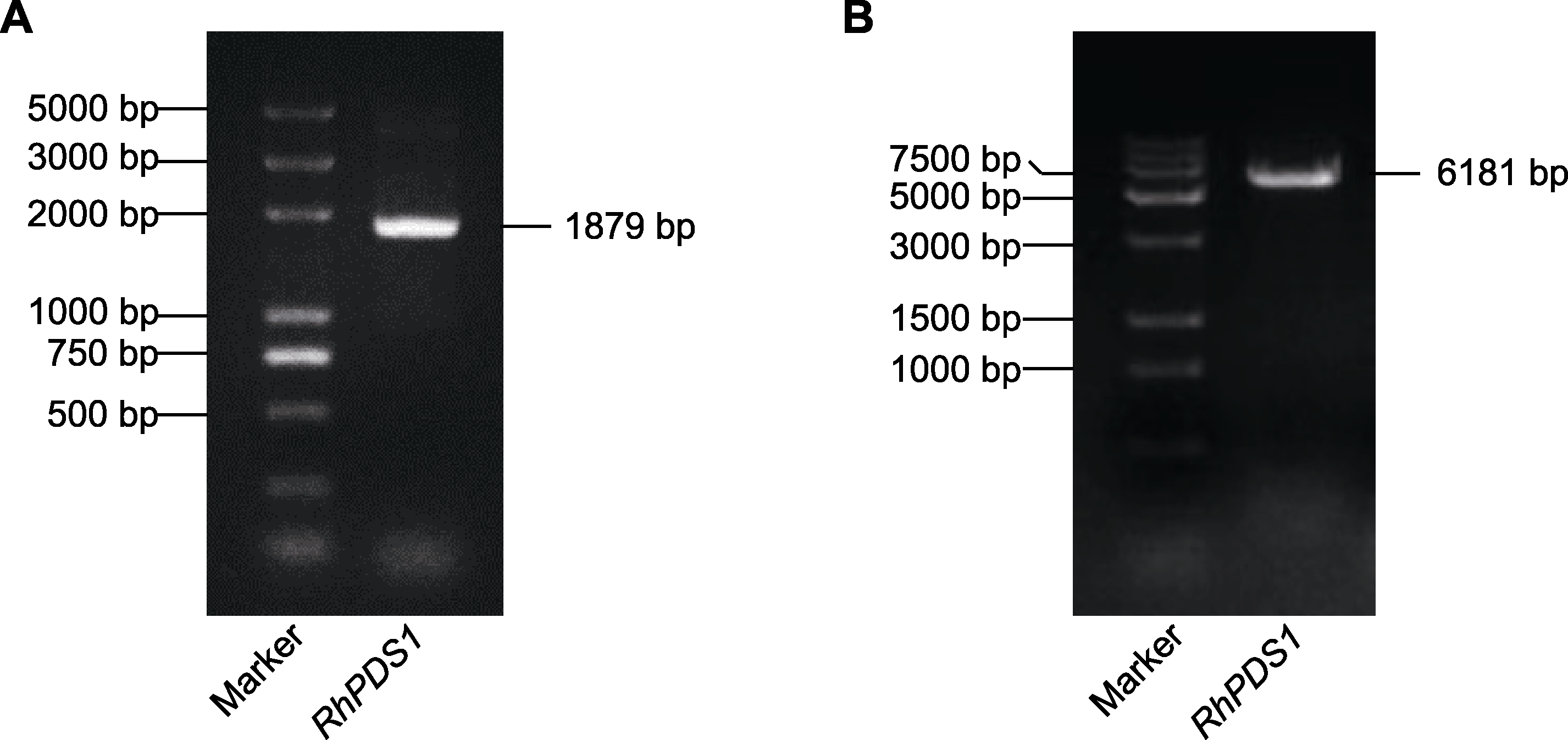
Figure 1 Electrophoresis detection of PCR amplified products of RhPDS1 gene (A) RhPDS1 was amplified with the cDNA of Rehmannia henryi leaves as a template; (B) RhPDS1 was amplified with the genomic DNA of R. henryi leaves as a template
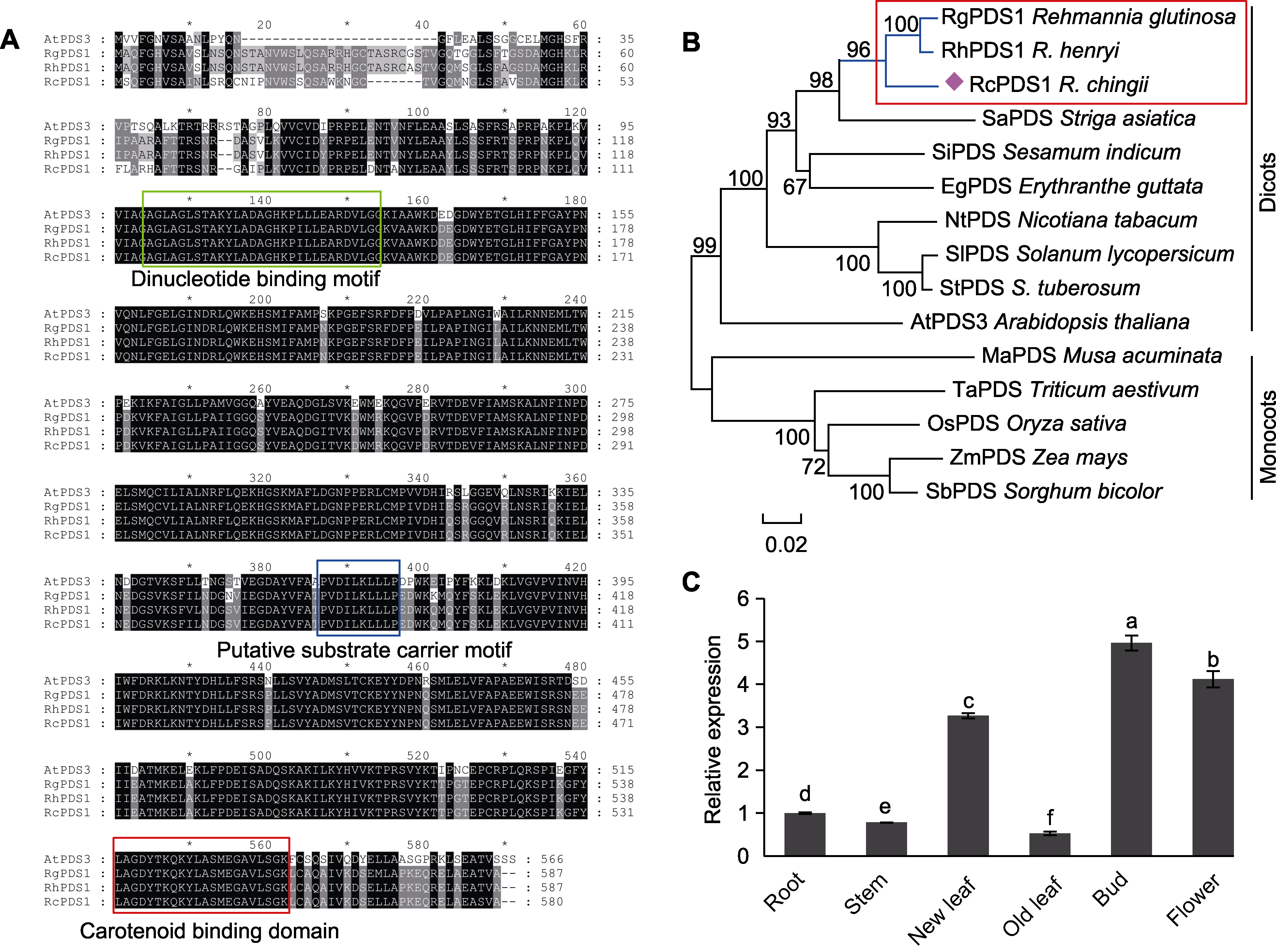
Figure 2 Sequence characteristics and gene expression patterns of RhPDS1 (A) Multiple sequence alignment of RhPDS1 with its homologs from Arabidopsis thaliana, Rehmannia glutinosa and R. chingii; (B) Phylogenetic tree of RhPDS1 and its homologous proteins; (C) Expression profiles of RhPDS1 in different tissues of R. henryi (different lowercase letters represent significant differences among different tissues (P<0.05)).
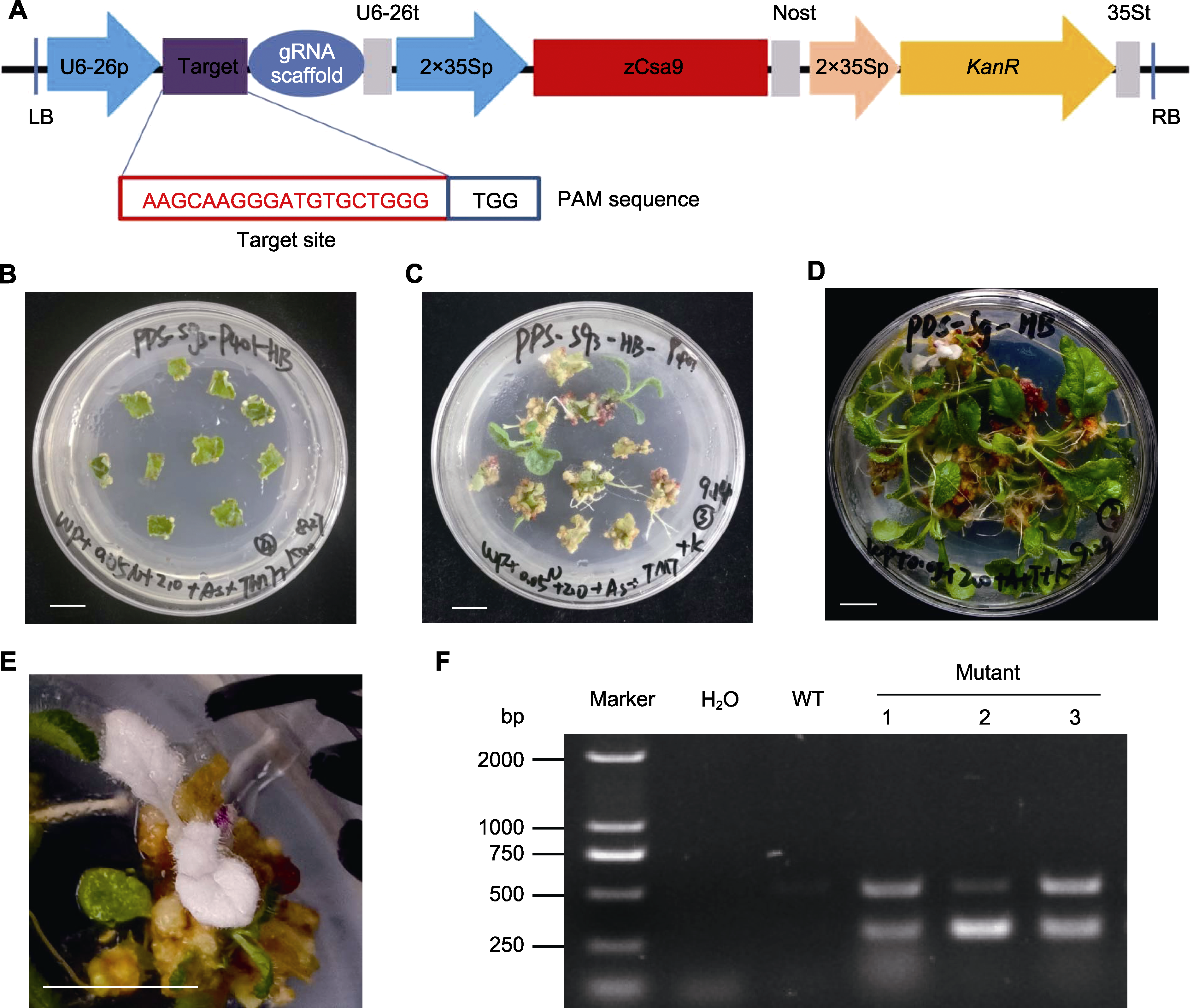
Figure 3 Construction of genome editing vector and genetic transformation of Rehmannia henryi (A) Schematic map of CRISPR/Cas9 construct for RhPDS1; (B) Embryogenic calli were screened on the medium with kanamycine (bar=1 cm); (C), (D) Resistant shoots differentiation (bars=1 cm); (E) Albino shoot regeneration (bar=1 cm); (F) Positive identification of albino shoots by PCR. PAM: Protospacer adjacent motif; WT: Wild type
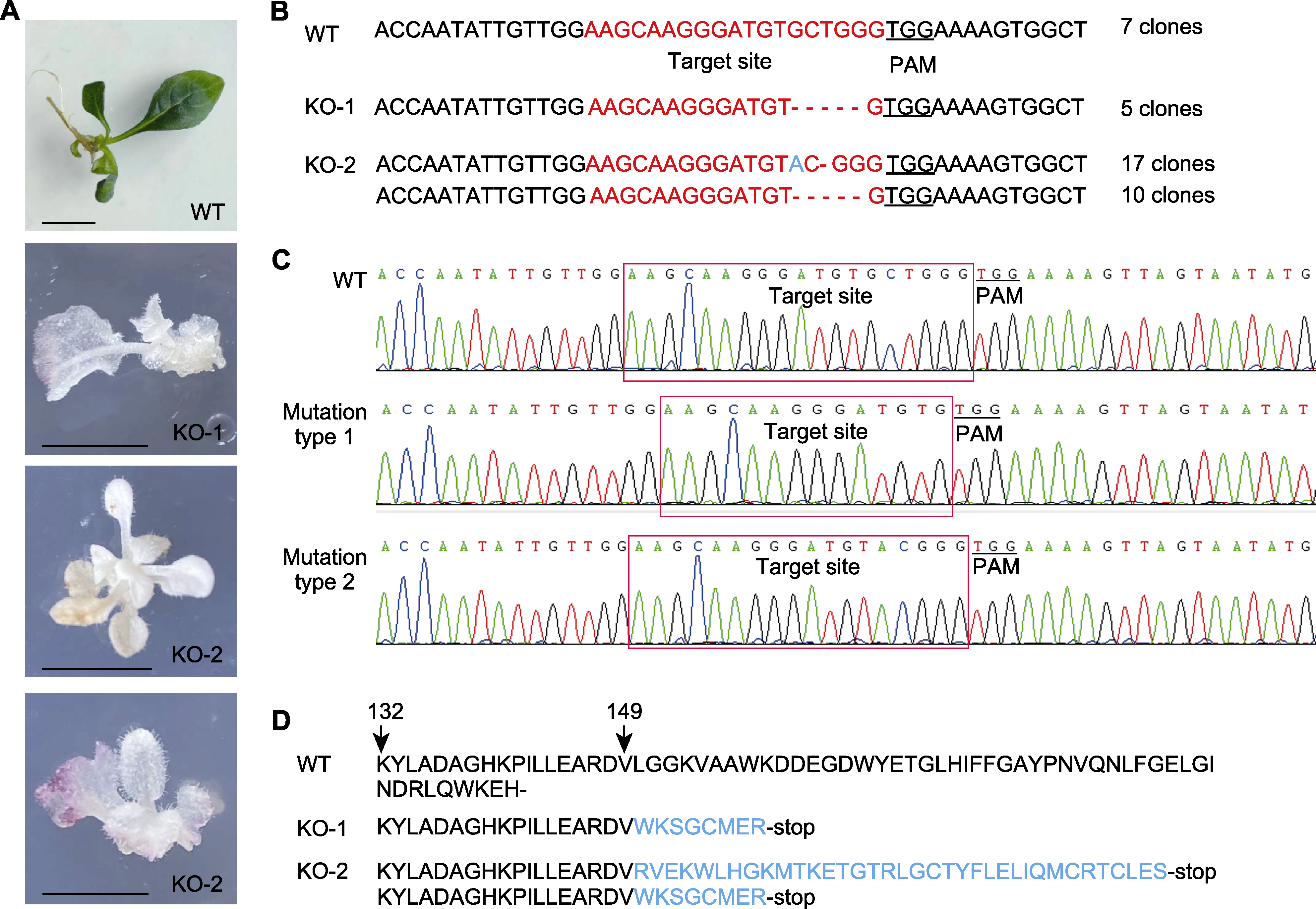
Figure 4 Analysis of target mutations of RhPDS1 gene in transgenic Rehmannia henryi (A) Phenotypes of the three albino shoots (bars=1 cm); (B) cDNA sequence characteristics of target site in the genome of two albino mutants (short horizontal lines represent base deletions); (C) Sanger sequencing results for T clone of two kinds of mutant; (D) Amino acids sequence characteristics of the two albino mutants. PAM and WT are the same as shown in Figure 3.
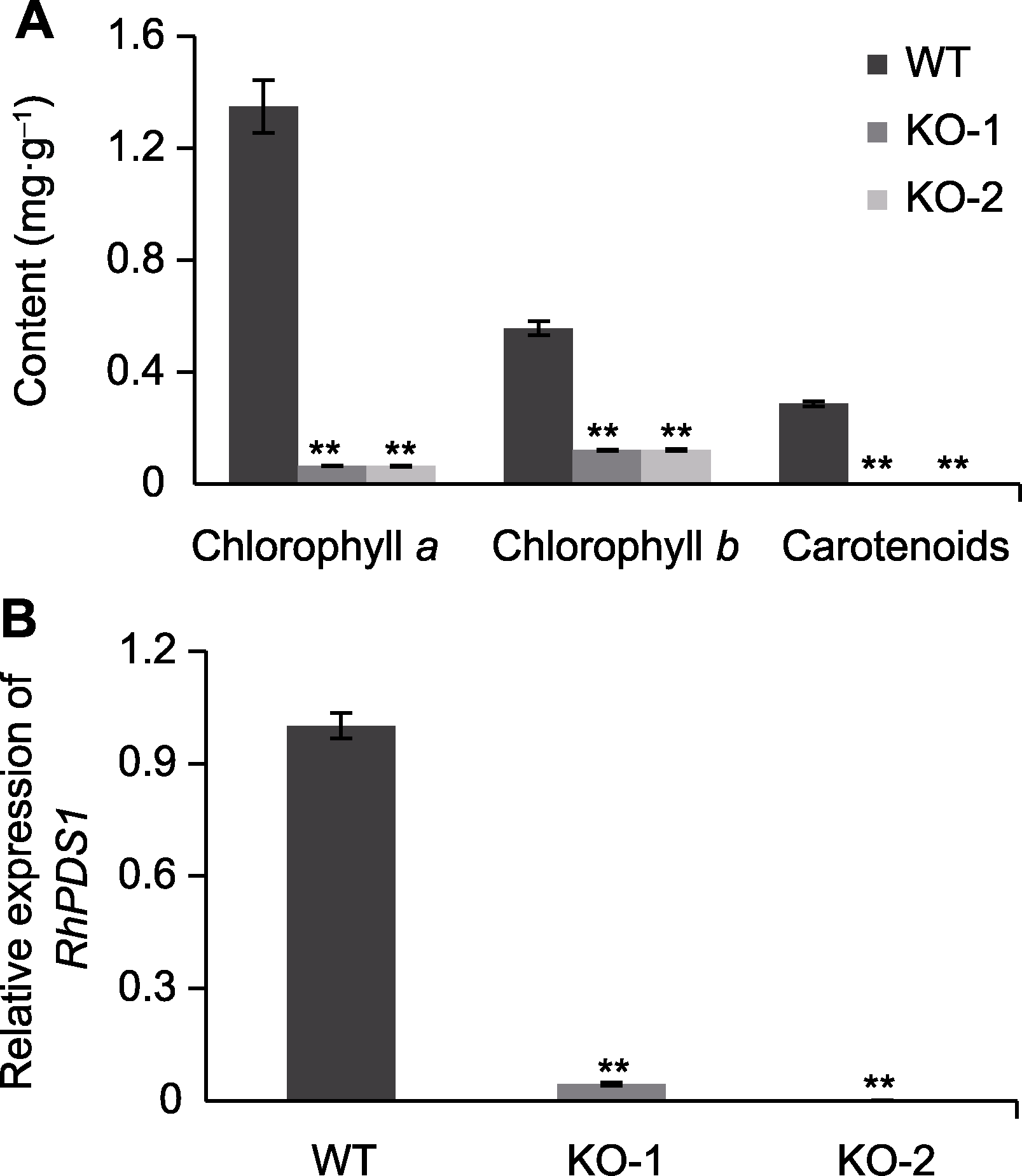
Figure 5 Detection of chlorophylls and carotenoids contents in RhPDS1 mutants and the expression of RhPDS1 (A) Chlorophylls and carotenoids contents in RhPDS1 mutants; (B) The relative expression levels of RhPDS1 in CRISPR/Cas9 edited shoots. Values are means±SD (n=3). ** indicate significant differences compared to wild type (WT) at P<0.01.
| [1] |
郭丽, 王雪涵, 田丰 (2023). 多组学整合网络: 一把精准解码玉米功能基因组的钥匙. 植物学报 58, 1-5.
DOI |
| [2] |
何晓玲, 刘鹏程, 马伯军, 陈析丰 (2022). 基于CRISPR/ Cas9的基因编辑技术研究进展及其在植物中的应用. 植物学报 57, 508-531.
DOI |
| [3] | 彭华胜, 徐长青, 袁媛, 查良平, 陈焕文, 管理, 康利平, 杨军, 王亚君, 曹丽娟, 程京, 黄璐琦 (2019). 最早的中药辅料炮制品: 西汉海昏侯墓出土的木质漆盒内样品鉴定与分析. 科学通报 64, 935-947. |
| [4] |
王丹, 王谧, 刘军, 周晓慧, 刘松瑜, 杨艳, 庄勇 (2022). 茄子U6启动子克隆及CRISPR/Cas9介导的基因编辑体系建立. 园艺学报 49, 791-800.
DOI |
| [5] | 吴琼, 孙超, 陈士林, 罗红梅, 李滢, 孙永珍, 牛云云 (2010). 转录组学在药用植物研究中的应用. 世界科学技术(中医药现代化) 12, 457-462. |
| [6] | 周婕 (2019). 湖北地黄化学成分研究. 硕士论文. 北京: 北京协和医学院. pp. 32-102. |
| [7] |
左鑫, 李铭铭, 李欣容, 苗春妍, 李炎枋, 杨旭, 张重义, 王丰青 (2022). CRISPR/Cas9技术在天目地黄RcPDS1基因编辑中的应用. 园艺学报 49, 1532-1544.
DOI |
| [8] |
Alagoz Y, Gurkok T, Zhang BH, Unver T (2016). Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in Opium poppy using CRISPR-Cas 9 genome editing technology. Sci Rep 6, 30910.
DOI PMID |
| [9] |
Banakar R, Schubert M, Collingwood M, Vakulskas C, Eggenberger AL, Wang K (2020). Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene. Rice 13, 4.
DOI PMID |
| [10] |
Bánfalvi Z, Csákvári E, Villányi V, Kondrák M (2020). Generation of transgene-free PDS mutants in potato by Agrobacterium-mediated transformation. BMC Biotechnol 20, 25.
DOI PMID |
| [11] |
Cheng JY, Hill C, Han Y, He TH, Ye XG, Shabala S, Guo GG, Zhou MX, Wang K, Li CD (2023). New semi-dwarfing alleles with increased coleoptile length by gene editing of gibberellin 3-oxidase 1 using CRISPR-Cas9 in barley (Hordeum vulgare L.). Plant Biotechnol J 21, 806-818.
DOI URL |
| [12] |
Kaur N, Alok A, Shivani, Kaur N, Pandey P, Awasthi P, Tiwari S (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct Integr Genomics 18, 89-99.
DOI URL |
| [13] |
Koschmieder J, Fehling-Kaschek M, Schaub P, Ghisla S, Brausemann A, Timmer J, Beyer P (2017). Plant-type phytoene desaturase: functional evaluation of structural implications. PLoS One 12, e0187628.
DOI URL |
| [14] | Kui L, Chen HT, Zhang WX, He SM, Xiong ZJ, Zhang YS, Yan L, Zhong CF, He FM, Chen JW, Zeng P, Zhang GH, Yang SC, Dong Y, Wang W, Cai J (2017). Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front Plant Sci 7, 2036. |
| [15] |
Li TD, Yang XP, Yu Y, Si XM, Zhai XW, Zhang HW, Dong WX, Gao CX, Xu C (2018). Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36, 1160-1163.
DOI |
| [16] |
Li XR, Zuo X, Li MM, Yang X, Zhi JY, Sun HZ, Xie CX, Zhang ZY, Wang FQ (2021). Efficient CRISPR/Cas9- mediated genome editing in Rehmannia glutinosa. Plant Cell Rep 40, 1695-1707.
DOI |
| [17] | Ma CF, Liu MC, Li QF, Si J, Ren XS, Song HY (2019). Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic Plant J 5, 164-169. |
| [18] |
Ma LG, Dong CM, Song C, Wang XL, Zheng XK, Niu Y, Chen SL, Feng WS (2021). De novo genome assembly of the potent medicinal plant Rehmannia glutinosa using nanopore technology. Comput Struct Biotechnol J 19, 3954-3963.
DOI URL |
| [19] |
Mao YF, Zhang ZJ, Feng ZY, Wei PL, Zhang H, Botella JR, Zhu JK (2016). Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14, 519-532.
DOI URL |
| [20] |
Odipio J, Alicai T, Ingelbrecht I, Nusinow DA, Bart R, Taylor NJ (2017). Efficient CRISPR/Cas9 genome editing of Phytoene desaturase in cassava. Front Plant Sci 8, 1780.
DOI PMID |
| [21] |
Qin GJ, Gu HY, Ma LG, Peng YB, Deng XW, Chen ZL, Qu LJ (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17, 471-482.
DOI |
| [22] |
Ren C, Liu YF, Guo YC, Duan W, Fan PG, Li SH, Liang ZC (2021). Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic Res 8, 52.
DOI |
| [23] |
Schmittgen TD, Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101-1108.
DOI PMID |
| [24] |
Son S, Park SR (2022). Challenges facing CRISPR/Cas9- based genome editing in plants. Front Plant Sci 13, 902413.
DOI URL |
| [25] |
Wang FQ, Li XR, Zuo X, Li MM, Miao CY, Zhi JY, Li YJ, Yang X, Liu XY, Xie CX (2021). Transcriptome-wide identification of WRKY transcription factor and functional characterization of RgWRKY37 involved in acteoside biosynthesis in Rehmannia glutinosa. Front Plant Sci 12, 739853.
DOI URL |
| [26] |
Wang J, Wu HT, Chen YN, Yin TM (2020). Efficient CRISPR/Cas9-mediated gene editing in an interspecific hybrid poplar with a highly heterozygous genome. Front Plant Sci 11, 996.
DOI PMID |
| [27] |
Wilson FM, Harrison K, Armitage AD, Simkin AJ, Harrison RJ (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45.
DOI PMID |
| [28] |
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14, 327.
DOI URL |
| [29] |
Xu ZS, Feng K, Xiong AS (2019). CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol Biotechnol 61, 191-199.
DOI |
| [30] |
Yu H, Lin T, Meng XB, Du HL, Zhang JK, Liu GF, Chen MJ, Jing YH, Kou LQ, Li XX, Gao Q, Liang Y, Liu XD, Fan ZL, Liang YT, Cheng ZK, Chen MS, Tian ZX, Wang YH, Chu CC, Zuo JR, Wan JM, Qian Q, Han B, Zuccolo A, Wing RA, Gao CX, Liang CZ, Li JY (2021). A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156-1170.
DOI URL |
| [31] |
Zhang YX, Malzahn AA, Sretenovic S, Qi YP (2019). The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants 5, 778-794.
DOI PMID |
| [32] |
Zhang YX, Xu GC, Cheng CH, Lei L, Sun J, Xu Y, Deng CH, Dai ZG, Yang ZM, Chen XJ, Liu C, Tang Q, Su JG (2021). Establishment of an Agrobacterium-mediated genetic transformation and CRISPR/Cas9-mediated targeted mutagenesis in hemp (Cannabis sativa L.). Plant Biotechnol J 19, 1979-1987.
DOI URL |
| [33] |
Zhou J, Shi GR, Liu YF, Chen RY, Yu DQ (2019). Nine new compounds from the whole plants of Rehmannia henryi. J Asian Nat Prod Res 21, 399-408.
DOI PMID |
| [34] |
Zhou Z, Tan HX, Li Q, Chen JF, Gao SH, Wang Y, Chen WS, Zhang L (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 148, 63-70.
DOI URL |
| [35] |
Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018). De novo domestication of wild tomato using genome editing. Nat Biotechnol 36, 1211-1216.
DOI |
| [36] | Zuo X, Wang FQ, Li XR, Li MM (2020). Transcriptome- based screening and the optimal reference genes for real-time quantitative PCR in Rehmannia chingii and R. henryi. Biol Plant 64, 798-806. |
| [1] | He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng. Advance in Gene-editing Technology Based on CRISPR/Cas9 and Its Application in Plants [J]. Chinese Bulletin of Botany, 2022, 57(4): 508-531. |
| [2] | Xianrong Xie, Dongchang Zeng, Jiantao Tan, Qinlong Zhu, Yaoguang Liu. CRISPR-based DNA Fragment Deletion in Plants [J]. Chinese Bulletin of Botany, 2021, 56(1): 44-49. |
| [3] | Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang. Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing [J]. Chinese Bulletin of Botany, 2019, 54(3): 385-395. |
| [4] | Wang Ying, Li Xianggan, Qiu Lijuan. Research Progress in Off-target in CRISPR/Cas9 Genome Editing [J]. Chinese Bulletin of Botany, 2018, 53(4): 528-541. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||