

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (3): 452-462.DOI: 10.11983/CBB23046 cstr: 32102.14.CBB23046
• SPECIAL TOPICS • Previous Articles Next Articles
Received:2023-04-07
Accepted:2023-08-07
Online:2024-05-10
Published:2024-05-10
Contact:
E-mail: Danling Hu, Yongwei Sun. Advances in Virus-mediated Genome Editing Technology in Plants[J]. Chinese Bulletin of Botany, 2024, 59(3): 452-462.
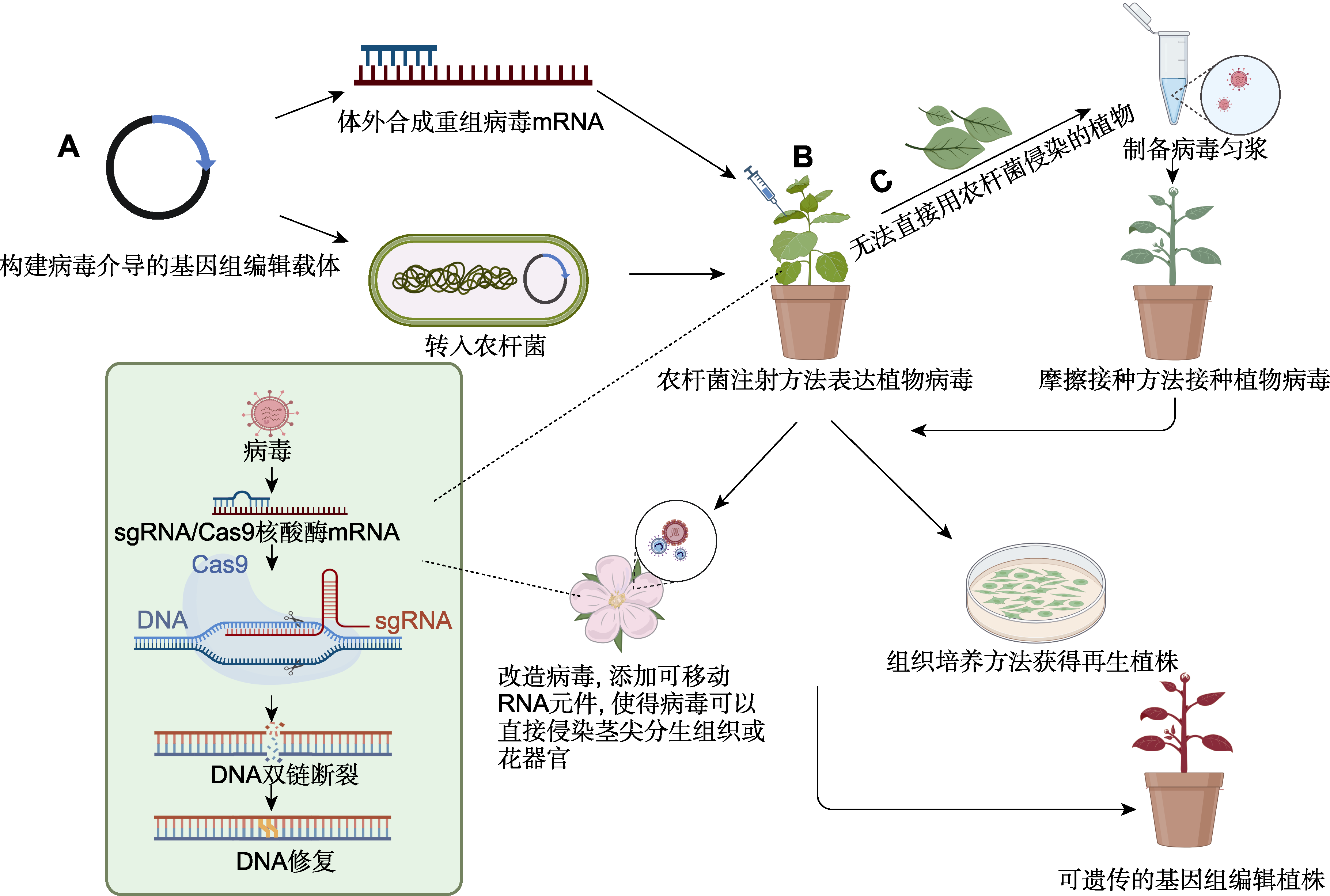
Figure 1 Flow chart of virus-mediated genome editing in plants (A) Engineering of viral vectors for transient expression of genome editing reagent; (B) Pathway of using Agrobacterium inoculation method to express plant virus for genome editing; (C) Pathway of delivery of virus to plants which cannot be infected by Agrobacterium inoculation
| 病毒类型 | 病毒名称 | 受体植物 | 递送成分 | 非组织培养稳定遗传效率 | 参考文献 |
|---|---|---|---|---|---|
| 正链RNA病毒 | 烟草脆裂病毒 | 本氏烟草(Nicotiana benthamiana) (Cas9过表达) | sgRNAs | 0.2% | Ali et al., |
| 烟草脆裂病毒和豌豆早枯病毒 | 本氏烟草和拟南芥(Arabidopsis thaliana) (Cas9过表达) | sgRNAs | N.A | Ali et al., | |
| 烟草脆裂病毒 | 本氏烟草(Cas9过表达) | sgRNAs和可移动sgRNAs | 65%-100% | Ellison et al., | |
| 烟草脆裂病毒 | 拟南芥(过表达SunTag系统) | sgRNAs和可移动 sgRNAs | 5%-8% | Ghoshal et al., | |
| 烟草脆裂病毒 | 拟南芥(Cas9过表达) | sgRNAs和可移动sgRNAs | 39%-60% | Nagalakshmi et al., | |
| 烟草脆裂病毒 | 烟草(N. tabacum) (Cas9过表达) | sgRNAs | N.A | Oh et al., | |
| 烟草脆裂病毒和豌豆早枯病毒 | 烟草(Cas9过表达) | sgRNAs | N.A | Kim et al., | |
| 烟草花叶病毒 | 本氏烟草(Cas9过表达) | Cas9和sgRNAs | N.A | Cody et al., | |
| 烟草花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Chiong et al., | |
| 番茄花叶病毒 | 本氏烟草(分开的SaCas9s部分过表达) | 部分Split-SaCas9s和sgRNAs | N.A | Kaya et al., | |
| 苹果潜隐球形病毒 | 本氏烟草和大豆(Glycine max) (Cas9, Csy4过表达) | sgRNAs | N.A | Luo et al., | |
| 甜菜坏死黄脉病毒 | 本氏烟草(Cas9过表达) | sgRNAs | N.A | Jiang et al., | |
| 大麦条纹花叶病毒 | 本氏烟草、小麦(Triticum aestivum)和玉米(Zea mays) (Cas9过表达) | sgRNAs | N.A | Hu et al., | |
| 大麦条纹花叶病毒 | 小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 0-17% | Wang et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 12.9%-100% | Li et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 0.5%-0.8% | Chen et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和大麦(Hordeum vulgare) (Cas9过表达) | sgRNAs | 100% | Tamilselvan-Nattar- Amutha et al., 2023 | |
| 马铃薯X病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Ariga et al., | |
| 马铃薯X病毒 | 本氏烟草(Cas9过表达) | sgRNAs和可移动sgRNAs | 22% | Uranga et al., | |
| 狗尾草花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Zhang et al., | |
| 狗尾草花叶病毒 | 本氏烟草、玉米和狗尾草(Setaria viridis) (Cas9过表达) | sgRNAs | N.A | Mei et al., | |
| 负链RNA 病毒 | 苦苣菜黄网弹状病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Ma et al., |
| 大麦黄条纹花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Gao et al., | |
| 番茄斑萎病毒 | 本氏烟草、番茄(Solanum lycopersicum)、辣椒(Capsicum annuum)、灯笼果(Physalis peruviana)和花生(Arachis hypogaea) | Cas9和sgRNAs, Cas12a和crRNA | N.A | Liu et al., | |
| DNA 病毒 | 甘蓝曲叶病毒 | 本氏烟草(Cas9过表达) | sgRNAs | N.A | Yin et al., |
| 棉皱叶病毒 | 拟南芥(Cas9过表达) | sgRNAs和可移动sgRNAs | 4.35%-8.79% | Lei et al., | |
| 棉皱叶病毒 | 棉花(Gossypium spp.) (Cas9过表达) | sgRNAs和可移动sgRNAs | N.A | Lei et al., |
Table 1 Virus-mediated CRISPR/Cas genome editing technology in plants
| 病毒类型 | 病毒名称 | 受体植物 | 递送成分 | 非组织培养稳定遗传效率 | 参考文献 |
|---|---|---|---|---|---|
| 正链RNA病毒 | 烟草脆裂病毒 | 本氏烟草(Nicotiana benthamiana) (Cas9过表达) | sgRNAs | 0.2% | Ali et al., |
| 烟草脆裂病毒和豌豆早枯病毒 | 本氏烟草和拟南芥(Arabidopsis thaliana) (Cas9过表达) | sgRNAs | N.A | Ali et al., | |
| 烟草脆裂病毒 | 本氏烟草(Cas9过表达) | sgRNAs和可移动sgRNAs | 65%-100% | Ellison et al., | |
| 烟草脆裂病毒 | 拟南芥(过表达SunTag系统) | sgRNAs和可移动 sgRNAs | 5%-8% | Ghoshal et al., | |
| 烟草脆裂病毒 | 拟南芥(Cas9过表达) | sgRNAs和可移动sgRNAs | 39%-60% | Nagalakshmi et al., | |
| 烟草脆裂病毒 | 烟草(N. tabacum) (Cas9过表达) | sgRNAs | N.A | Oh et al., | |
| 烟草脆裂病毒和豌豆早枯病毒 | 烟草(Cas9过表达) | sgRNAs | N.A | Kim et al., | |
| 烟草花叶病毒 | 本氏烟草(Cas9过表达) | Cas9和sgRNAs | N.A | Cody et al., | |
| 烟草花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Chiong et al., | |
| 番茄花叶病毒 | 本氏烟草(分开的SaCas9s部分过表达) | 部分Split-SaCas9s和sgRNAs | N.A | Kaya et al., | |
| 苹果潜隐球形病毒 | 本氏烟草和大豆(Glycine max) (Cas9, Csy4过表达) | sgRNAs | N.A | Luo et al., | |
| 甜菜坏死黄脉病毒 | 本氏烟草(Cas9过表达) | sgRNAs | N.A | Jiang et al., | |
| 大麦条纹花叶病毒 | 本氏烟草、小麦(Triticum aestivum)和玉米(Zea mays) (Cas9过表达) | sgRNAs | N.A | Hu et al., | |
| 大麦条纹花叶病毒 | 小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 0-17% | Wang et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 12.9%-100% | Li et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和小麦(Cas9过表达) | sgRNAs和可移动sgRNAs | 0.5%-0.8% | Chen et al., | |
| 大麦条纹花叶病毒 | 本氏烟草和大麦(Hordeum vulgare) (Cas9过表达) | sgRNAs | 100% | Tamilselvan-Nattar- Amutha et al., 2023 | |
| 马铃薯X病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Ariga et al., | |
| 马铃薯X病毒 | 本氏烟草(Cas9过表达) | sgRNAs和可移动sgRNAs | 22% | Uranga et al., | |
| 狗尾草花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Zhang et al., | |
| 狗尾草花叶病毒 | 本氏烟草、玉米和狗尾草(Setaria viridis) (Cas9过表达) | sgRNAs | N.A | Mei et al., | |
| 负链RNA 病毒 | 苦苣菜黄网弹状病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Ma et al., |
| 大麦黄条纹花叶病毒 | 本氏烟草 | Cas9和sgRNAs | N.A | Gao et al., | |
| 番茄斑萎病毒 | 本氏烟草、番茄(Solanum lycopersicum)、辣椒(Capsicum annuum)、灯笼果(Physalis peruviana)和花生(Arachis hypogaea) | Cas9和sgRNAs, Cas12a和crRNA | N.A | Liu et al., | |
| DNA 病毒 | 甘蓝曲叶病毒 | 本氏烟草(Cas9过表达) | sgRNAs | N.A | Yin et al., |
| 棉皱叶病毒 | 拟南芥(Cas9过表达) | sgRNAs和可移动sgRNAs | 4.35%-8.79% | Lei et al., | |
| 棉皱叶病毒 | 棉花(Gossypium spp.) (Cas9过表达) | sgRNAs和可移动sgRNAs | N.A | Lei et al., |
| [1] | Ali Z, Abul-Faraj A, Li LX, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, Dinesh- Kumar S, Mahfouz MM (2015a). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant 8, 1288-1291. |
| [2] | Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM (2015b). Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav 10, e1044191. |
| [3] | Ali Z, Eid A, Ali S, Mahfouz MM (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res 244, 333-337. |
| [4] |
Al-Shayeb B, Skopintsev P, Soczek KM, Stahl EC, Li Z, Groover E, Smock D, Eggers AR, Pausch P, Cress BF, Huang CJ, Staskawicz B, Savage DF, Jacobsen SE, Banfield JF, Doudna JA (2022). Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell 185, 4574-4586.
DOI PMID |
| [5] | Ariga H, Toki S, Ishibashi K (2020). Potato virus X vector- mediated DNA-free genome editing in plants. Plant Cell Physiol 61, 1946-1953. |
| [6] |
Avesani L, Marconi G, Morandini F, Albertini E, Bruschet-ta M, Bortesi L, Pezzotti M, Porceddu A (2007). Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Res 16, 587-597.
PMID |
| [7] |
Barrangou R (2014). Cas9 targeting and the CRISPR revolution. Science 344, 707-708.
DOI PMID |
| [8] |
Bernabé-Orts JM, Casas-Rodrigo I, Minguet EG, Landolfi V, Garcia-Carpintero V, Gianoglio S, Vázquez-Vilar M, Granell A, Orzaez D (2019). Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol J 17, 1971-1984.
DOI PMID |
| [9] |
Bigelyte G, Young JK, Karvelis T, Budre K, Zedaveinyte R, Djukanovic V, Van Ginkel E, Paulraj S, Gasior S, Jones S, Feigenbutz L, Clair GS, Barone P, Bohn J, Acharya A, Zastrow-Hayes G, Henkel-Heinecke S, Silanskas A, Seidel R, Siksnys V (2021). Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modifi-cation in cells. Nat Commun 12, 6191.
DOI PMID |
| [10] | Cai QN, Guo DM, Cao YJ, Li Y, Ma R, Liu WP (2022). App- lication of CRISPR/CasΦ2 system for genome editing in plants. Int J Mol Sci 23, 5755. |
| [11] | Cao XS, Xie HT, Song ML, Lu JH, Ma P, Huang BY, Wang MG, Tian YF, Chen F, Peng J, Lang ZB, Li GF, Zhu JK (2022). Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation (Camb) 4, 100345. |
| [12] |
Chapman S, Faulkner C, Kaiserli E, Garcia-Mata C, Savenkov EI, Roberts AG, Oparka KJ, Christie JM (2008). The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci USA 105, 20038-20043.
DOI PMID |
| [13] | Chen H, Su ZQ, Tian B, Liu Y, Pang YH, Kavetskyi V, Trick HN, Bai GH (2022). Development and optimization of a Barley stripe mosaic virus-mediated gene editing system to improve Fusarium head blight resistance in wheat. Plant Biotechnol J 20, 1018-1020. |
| [14] |
Chen KL, Wang YP, Zhang R, Zhang HW, Gao CX (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70, 667-697.
DOI PMID |
| [15] | Chiong KT, Cody WB, Scholthof HB (2021). RNA silencing suppressor-influenced performance of a virus vector de- livering both guide RNA and Cas9 for CRISPR gene editing. Sci Rep 11, 6769. |
| [16] | Cody WB, Scholthof HB, Mirkov TE (2017). Multiplexed gene editing and protein overexpression using a Tobacco mosaic virus viral vector. Plant Physiol 175, 23-35. |
| [17] |
Creager ANH, Scholthof KBG, Citovsky V, Scholthof HB (1999). Tobacco mosaic virus: pioneering research for a century. Plant Cell 11, 301-308.
PMID |
| [18] |
Ellison EE, Nagalakshmi U, Gamo ME, Huang PJ, Dinesh-Kumar S, Voytas DF (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants 6, 620-624.
DOI PMID |
| [19] |
Ganesan U, Bragg JN, Deng M, Marr S, Lee MY, Qian SS, Shi ML, Kappel J, Peters C, Lee Y, Goodin MM, Dietzgen RG, Li ZH, Jackson AO (2013). Construction of a Sonchus yellow net virus minireplicon: a step toward re- verse genetic analysis of plant negative-strand RNA viru- ses. J Virol 87, 10598-10611.
DOI PMID |
| [20] | Gao CX (2018). The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol 19, 275-276. |
| [21] | Gao Q, Xu WY, Yan T, Fang XD, Cao Q, Zhang ZJ, Ding ZH, Wang Y, Wang XB (2019). Rescue of a plant cytorhabdo virus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol 223, 2120-2133. |
| [22] | Ghoshal B, Vong B, Picard CL, Feng SH, Tam JM, Jacobsen SE (2020). A viral guide RNA delivery system for CRISPR-based transcriptional activation and heritable targeted DNA demethylation in Arabidopsis thaliana. PLo- S Genet 16, e1008983. |
| [23] | He XL, Liu PC, Ma BJ, Chen XF (2022). Advance in gene- editing technology based on CRISPR/Cas9 and its appli- cation in plants. Chin Bull Bot 57, 508-531. (in Chinese) |
|
何晓玲, 刘鹏程, 马伯军, 陈析丰 (2022). 基于CRISPR/Cas9的基因编辑技术研究进展及其在植物中的应用. 植物学报 57, 508-531.
DOI |
|
| [24] | He YB, Zhang T, Sun H, Zhan HD, Zhao YD (2020). A reporter for noninvasively monitoring gene expression and plant transformation. Hortic Res 7, 152. |
| [25] |
Hu JC, Li SY, Li ZL, Li HY, Song WB, Zhao HM, Lai JS, Xia LQ, Li DW, Zhang YL (2019). A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol Plant Pathol 20, 1463-1474.
DOI PMID |
| [26] |
Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M (2005). Biology of plant rhabdoviruses. Annu Rev Phytopathol 43, 623-660.
PMID |
| [27] |
Jiang N, Zhang C, Liu JY, Guo ZH, Zhang ZY, Han CG, Wang Y (2019). Development of Beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol J 17, 1302-1315.
DOI PMID |
| [28] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
DOI PMID |
| [29] | Kaya H, Ishibashi K, Toki S (2017). A split Staphylococcus aureus Cas9 as a compact genome-editing tool in plants. Plant Cell Physiol 58, 643-649. |
| [30] | Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, Koo T, Ko JH, Kim YS (2022). Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol 40, 94-102. |
| [31] | Kim H, Oh Y, Park E, Kang M, Choi Y, Kim SG (2023). Heritable virus-induced genome editing (VIGE) in Nicotiana attenuata. In: Nicotiana attenuata. In: Bae S, Song B, eds. Base Editors: Methods and Protocols. New York: Humana. pp. 203-218. |
| [32] | Lee WS, Hammond-Kosack KE, Kanyuka K (2012). Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus- mediated overexpression of heterologous protein. Plant Physiol 160, 582-590. |
| [33] | Lei JF, Dai PH, Li Y, Zhang WQ, Zhou GT, Liu C, Liu XD (2021). Heritable gene editing using FT mobile guide RNAs and DNA viruses. Plant Methods 17, 20. |
| [34] | Lei JF, Li Y, Dai PH, Liu C, Zhao Y, You YZ, Qu YY, Chen QJ, Liu XD (2022). Efficient virus-mediated genome editing in cotton using the CRISPR/Cas9 system. Front Plant Sci 13, 1032799. |
| [35] |
Li SY, Li JY, He YB, Xu ML, Zhang JH, Du WM, Zhao YD, Xia LQ (2019). Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol 37, 445-450.
DOI PMID |
| [36] |
Li SY, Xia LQ (2020). Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives. aBIOTECH 1, 58-73.
DOI PMID |
| [37] |
Li TD, Hu JC, Sun Y, Li BS, Zhang DL, Li WL, Liu JX, Li DW, Gao CX, Zhang YL, Wang YP (2021). Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol Plant 14, 1787-1798.
DOI PMID |
| [38] | Li Z, Zhong ZH, Wu ZS, Pausch P, Al-Shayeb B, Amerasekera J, Doudna JA, Jacobsen SE (2023). Genome editing in plants using the compact editor casΦ. Proc Natl Acad Sci USA 120, e2216822120. |
| [39] |
Liang Z, Chen KL, Li TD, Zhang Y, Wang YP, Zhao Q, Liu JX, Zhang HW, Liu CM, Ran YD, Gao CX (2017). Effi- cient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8, 14261.
DOI PMID |
| [40] | Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HCW, Zhang YS, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16, 1295-1310. |
| [41] |
Liu HW, Zhang BH (2020). Virus-based CRISPR/Cas9 genome editing in plants. Trends Genet 36, 810-813.
DOI PMID |
| [42] | Liu Q, Zhao CL, Sun K, Deng YL, Li ZH (2023). Engineered biocontainable RNA virus vectors for non-transgenic ge- nome editing across crop species and genotypes. Mol Plant 16, 616-631. |
| [43] | Liu SS, Sretenovic S, Fan TT, Cheng YH, Li G, Qi A, Tang X, Xu Y, Guo WJ, Zhong ZH, He Y, Liang YL, Han QQ, Zheng XL, Gu XF, Qi YP, Zhang Y (2022). Hypercompact CRISPR-Cas12j2 (CasΦ) enables genome editing, gene activation, and epigenome editing in plants. Plant Commun 3, 100453. |
| [44] | Liu YG, Li GS, Zhang YL, Chen LT (2019). Current advances on CRISPR/Cas genome editing technologies in plants. J South China Agricul Univ 40(5), 38-49. (in Chinese) |
| 刘耀光, 李构思, 张雅玲, 陈乐天 (2019). CRISPR/Cas植物基因组编辑技术研究进展. 华南农业大学学报 40(5), 38-49. | |
| [45] |
Luo YJ, Na R, Nowak JS, Qiu Y, Lu QS, Yang CY, Marsolais F, Tian LN (2021). Development of a Csy4-processed guide RNA delivery system with soybean-infecting virus ALSV for genome editing. BMC Plant Biol 21, 419.
DOI PMID |
| [46] |
Ma XN, Zhang XY, Liu HM, Li ZH (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6, 773-779.
DOI PMID |
| [47] | Mei Y, Beernink BM, Ellison EE, Konečná E, Neelakandan AK, Voytas DF, Whitham SA (2019). Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 3, e00181. |
| [48] |
Nagalakshmi U, Meier N, Liu JY, Voytas DF, Dinesh-Kumar SP (2022). High-efficiency multiplex biallelic heritable editing in Arabidopsis using an RNA virus. Plant Physiol 189, 1241-1245.
DOI PMID |
| [49] | Oh Y, Kim H, Lee HJ, Kim SG (2022). Ribozyme-processed guide RNA enhances virus-mediated plant genome editing. Biotechnol J 17, 2100189. |
| [50] |
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA (2020). CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333-337.
DOI PMID |
| [51] | Purkayastha A, Dasgupta I (2009). Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol Biochem 47, 967-976. |
| [52] |
Senthil-Kumar M, Mysore KS (2011). New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16, 656-665.
DOI PMID |
| [53] | Su YK, Qiu JR, Zhang H, Song ZQ, Wang JH (2019). Recent progress in evolutionary technology of CRISPR/Cas9 system for plant genome editing. Chin Bull Bot 54, 385-395. (in Chinese) |
|
苏钺凯, 邱镜仁, 张晗, 宋振巧, 王建华 (2019). CRISPR/ Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报 54, 385-395.
DOI |
|
| [54] | Tai YS, Bragg J, Meinhardt SW (2007). Functional characterization of toxa and molecular identification of its intracellular targeting protein in wheat. Am J Plant Physiol 2, 76-89. |
| [55] | Tamilselvan-Nattar-Amutha S, Hiekel S, Hartmann F, Lorenz J, Dabhi RV, Dreissig S, Hensel G, Kumlehn J, Heckmann S (2023). Barley stripe mosaic virus-mediated somatic and heritable gene editing in barley (Hordeum vulgare L.). Front Plant Sci 14, 1201446. |
| [56] | Uranga M, Aragonés V, Selma S, Vázquez-Vilar M, Orzáez D, Daròs JA (2021). Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J 106, 555-565. |
| [57] |
Wang D, Tai PWL, Gao GP (2019). Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18, 358-378.
DOI PMID |
| [58] | Wang Q, Ma XN, Qian SS, Zhou X, Sun K, Chen XL, Zhou XP, Jackson AO, Li ZH (2015). Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog 11, e1005223. |
| [59] |
Wang W, Yu ZT, He F, Bai GH, Trick HN, Akhunova A, Akhunov E (2022). Multiplexed promoter and gene editing in wheat using a virus-based guide RNA delivery system. Plant Biotechnol J 20, 2332-2341.
DOI PMID |
| [60] |
Wu ZW, Zhang YF, Yu HP, Pan D, Wang YJ, Wang YN, Li F, Liu C, Nan H, Chen WZ, Ji QJ (2021). Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol 17, 1132-1138.
DOI PMID |
| [61] |
Yin KQ, Han T, Liu G, Chen TY, Wang Y, Yu AYL, Liu YL (2015). A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5, 14926.
DOI PMID |
| [62] | Yip BH (2020). Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules 10, 839. |
| [63] | Yuan C, Li C, Yan LJ, Jackson AO, Liu ZY, Han CG, Yu JL, Li DW (2011). A high throughput Barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6, e26468. |
| [64] |
Zhan XQ, Lu YM, Zhu JK, Botella JR (2021). Genome editing for plant research and crop improvement. J Integr Plant Biol 63, 3-33.
DOI |
| [65] | Zhang X, Kang LH, Zhang Q, Meng QQ, Pan YF, Yu ZM, Shi NN, Jackson S, Zhang XL, Wang HZ, Tor M, Hong YG (2020). An RNAi suppressor activates in planta virus- mediated gene editing. Funct Integr Genomics 20, 471-477. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
