

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (1): 119-132.DOI: 10.11983/CBB18047 cstr: 32102.14.CBB18047
• SPECIAL TOPICS • Previous Articles Next Articles
Tong Qin1,2,Zhen Huang2,*( ),Zhenhui Kang1,*(
),Zhenhui Kang1,*( )
)
Received:2018-02-17
Accepted:2018-05-23
Online:2019-01-01
Published:2019-07-31
Contact:
Zhen Huang,Zhenhui Kang
Tong Qin,Zhen Huang,Zhenhui Kang. Regulatory Mechanism of Thioredoxin (Trx) in Chloroplasts[J]. Chinese Bulletin of Botany, 2019, 54(1): 119-132.
| 名称 | 登录号 | 氧化还原电位* | 功能 | 参考文献 |
|---|---|---|---|---|
| FTRA1 | AT5G23440 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| FTRA2 | AT5G08410 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| FTRB | AT2G04700 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| NTRC | AT2G41680 | -330 mV | 还原酶 | Michalska et al., 2009; Chae et al., 2013; Bernal-Bayard et al., 2014; Puerto-Galán et al., 2015; Carrilloet al., 2016; Naranjo et al., 2016b; Pérez-Ruiz et al., 2017 |
| Trx f1 | AT3G02730 | -321 mV (pH7.5) | 调节FBPase和ATP合酶(CF 1 γ亚基)活性, 氧化胁迫 | Hisabori et al., 2013; Yoshida et al., 2015 |
| Trx f2 | AT5G16400 | -321 mV (pH7.5) | 调节FBPase和ATP合酶(CF 1 γ亚基)活性, 氧化胁迫 | Hisabori et al., 2013; Yoshida et al., 2015 |
| Trx m1 | AT1G03680 | -335 mV (pH7.5) | 调节Calvin-Benson循环酶活性 | Okegawa and Motohashi, 2015 |
| Trx m2 | AT4G03520 | -335 mV (pH7.5) | 调节Calvin-Benson循环酶活性 | Okegawa and Motohashi, 2015 |
| Trx m3 | AT2G15570 | -316 mV (pH7.5) | 胞内蛋白运输和分生组织维持 | Benitez-Alfonso et al., 2009 |
| Trx m4 | AT3G15360 | -312 mV (pH7.5) | 调节Calvin-Benson循环酶活性, 环形电子传递 | Okegawa and Motohashi, 2015 |
| Trx y1 | AT1G76760 | -296 mV (pH7.5) | 胁迫响应 | Okegawa and Motohashi, 2015 |
| Trx y2 | AT1G43560 | -295 mV (pH7.5) | 硫代谢 | Laugier et al., 2013 |
| Trx x | AT1G50320 | -310 mV (pH7.5) | 胁迫响应 | Bernal-Bayard et al., 2014 |
| Trx z | AT3G06730 | -276 mV (pH7.5) | 质体转录胁迫应激 | Arsova et al., 2010; Chibani et al., 2011; Díaz et al., 2018 |
| HCF164 | At4G37200 | -224 mV ** | Cyt b6f复合体装配 | Motohashi and Hisabori, 2010 |
| LTO1 | AT4G35760 | -180 mV *** | 蛋白折叠 | Wang et al., 2011; Karamoko et al., 2011, 2013 |
| SOQ1 | At1G56500 | 未知 | 光抑制淬灭 | Brooks et al., 2013 |
| CDSP32 | AT1G76080 | -337 mV (pH7.9) | 调节MSRB1的活性 | Tarrago et al., 2010 |
| AtACHT1 (Lilium 5) | AT4G26160 | -237 mV | 抗氧化 | Dangoor et al., 2009, 2012 |
| AtACHT2a (Lilium 2) | AT4G29670.1 | -239 mV | 未知 | Dangoor et al., 2009 |
| AtACHT2b (Lilium 2) | AT4G29670.2 | 未知 | 未知 | Dangoor et al., 2009 |
| AtACHT3 (Lilium 4) | AT2G33270 | 未知 | 未知 | Dangoor et al., 2009 |
| AtACHT4a (Lilium 1) | AT1G08570.1 | -240 mV | 淀粉合成 | Dangoor et al., 2009; Eliyahu et al., 2015 |
| AtACHT4b (Lilium 1) | AT1G08570.2 | 未知 | 淀粉合成 | Dangoor et al., 2009; Eliyahu et al., 2015 |
| AtACHT5 (Lilium 3) | AT5G61440 | 未知 | 未知 | Dangoor et al., 2009 |
| WCRKC1 | AT5G06690 | 未知 | 冷胁迫 | Chibani et al., 2011 |
| WCRKC2 | AT5G04260 | 未知 | 冷胁迫 | Chibani et al., 2011 |
Table 1 Chloroplast Trx systems in Arabidopsis
| 名称 | 登录号 | 氧化还原电位* | 功能 | 参考文献 |
|---|---|---|---|---|
| FTRA1 | AT5G23440 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| FTRA2 | AT5G08410 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| FTRB | AT2G04700 | -356 mV | 还原酶 | Balsera et al., 2013; Wang et al., 2014; Yoshida and Hisabori, 2017 |
| NTRC | AT2G41680 | -330 mV | 还原酶 | Michalska et al., 2009; Chae et al., 2013; Bernal-Bayard et al., 2014; Puerto-Galán et al., 2015; Carrilloet al., 2016; Naranjo et al., 2016b; Pérez-Ruiz et al., 2017 |
| Trx f1 | AT3G02730 | -321 mV (pH7.5) | 调节FBPase和ATP合酶(CF 1 γ亚基)活性, 氧化胁迫 | Hisabori et al., 2013; Yoshida et al., 2015 |
| Trx f2 | AT5G16400 | -321 mV (pH7.5) | 调节FBPase和ATP合酶(CF 1 γ亚基)活性, 氧化胁迫 | Hisabori et al., 2013; Yoshida et al., 2015 |
| Trx m1 | AT1G03680 | -335 mV (pH7.5) | 调节Calvin-Benson循环酶活性 | Okegawa and Motohashi, 2015 |
| Trx m2 | AT4G03520 | -335 mV (pH7.5) | 调节Calvin-Benson循环酶活性 | Okegawa and Motohashi, 2015 |
| Trx m3 | AT2G15570 | -316 mV (pH7.5) | 胞内蛋白运输和分生组织维持 | Benitez-Alfonso et al., 2009 |
| Trx m4 | AT3G15360 | -312 mV (pH7.5) | 调节Calvin-Benson循环酶活性, 环形电子传递 | Okegawa and Motohashi, 2015 |
| Trx y1 | AT1G76760 | -296 mV (pH7.5) | 胁迫响应 | Okegawa and Motohashi, 2015 |
| Trx y2 | AT1G43560 | -295 mV (pH7.5) | 硫代谢 | Laugier et al., 2013 |
| Trx x | AT1G50320 | -310 mV (pH7.5) | 胁迫响应 | Bernal-Bayard et al., 2014 |
| Trx z | AT3G06730 | -276 mV (pH7.5) | 质体转录胁迫应激 | Arsova et al., 2010; Chibani et al., 2011; Díaz et al., 2018 |
| HCF164 | At4G37200 | -224 mV ** | Cyt b6f复合体装配 | Motohashi and Hisabori, 2010 |
| LTO1 | AT4G35760 | -180 mV *** | 蛋白折叠 | Wang et al., 2011; Karamoko et al., 2011, 2013 |
| SOQ1 | At1G56500 | 未知 | 光抑制淬灭 | Brooks et al., 2013 |
| CDSP32 | AT1G76080 | -337 mV (pH7.9) | 调节MSRB1的活性 | Tarrago et al., 2010 |
| AtACHT1 (Lilium 5) | AT4G26160 | -237 mV | 抗氧化 | Dangoor et al., 2009, 2012 |
| AtACHT2a (Lilium 2) | AT4G29670.1 | -239 mV | 未知 | Dangoor et al., 2009 |
| AtACHT2b (Lilium 2) | AT4G29670.2 | 未知 | 未知 | Dangoor et al., 2009 |
| AtACHT3 (Lilium 4) | AT2G33270 | 未知 | 未知 | Dangoor et al., 2009 |
| AtACHT4a (Lilium 1) | AT1G08570.1 | -240 mV | 淀粉合成 | Dangoor et al., 2009; Eliyahu et al., 2015 |
| AtACHT4b (Lilium 1) | AT1G08570.2 | 未知 | 淀粉合成 | Dangoor et al., 2009; Eliyahu et al., 2015 |
| AtACHT5 (Lilium 3) | AT5G61440 | 未知 | 未知 | Dangoor et al., 2009 |
| WCRKC1 | AT5G06690 | 未知 | 冷胁迫 | Chibani et al., 2011 |
| WCRKC2 | AT5G04260 | 未知 | 冷胁迫 | Chibani et al., 2011 |
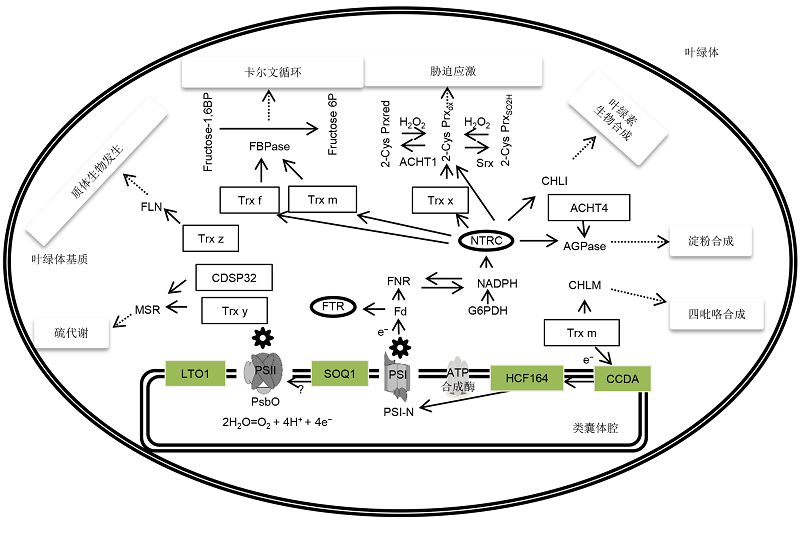
Figure 1 Chloroplast Trx systems and their functions The Trx systems in chloroplast play important roles in regulating key enzymes of Calvin cycle, plastid biogenesis, sulfur metabolism, stress response, and the synthesis of chlorophyll, tetrapyrrole, and starch. Since FTR affects all 5 Trx proteins, the lines are not marked in the figure. The target of SOQ1 on the PSII subunits is still unknown.
| 1 |
孙虎, 薛保国, 杨丽荣, 全鑫, 朱自贤, 武超, 马雪娇 ( 2010). 植物硫氧还蛋白系统. 基因组学与应用生物学 29, 748-753.
DOI URL |
| 2 |
张艳玲, 孙旭武, 张立新 ( 2009). 拟南芥叶绿体中DEG蛋白酶功能的研究进展. 植物学报 44, 37-42.
DOI URL |
| 3 | Arnoux P, Morosinotto T, Saga G, Bassi R, Pignol D ( 2009). A structural basis for the pH-dependent xanthophyll cycle in Arabidopsis thaliana.Plant Cell 21, 2036-2044. |
| 4 |
Arsova B, Hoja U, Wimmelbacher M, Greiner E, Üstün S, Melzer M, Petersen K, Lein W, Börnke F ( 2010). Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana.Plant Cell 22, 1498-1515.
DOI URL PMID |
| 5 |
Balsera M, Uberegui E, Schürmann P, Buchanan BB ( 2014). Evolutionary development of redox regulation in chloroplasts. Antioxid Redox Signal 21, 1327-1355.
DOI URL PMID |
| 6 |
Balsera M, Uberegui E, Susanti D, Schmitz RA, Mukhopadhyay B, Schürmann P, Buchanan BB ( 2013). Ferredoxin: thioredoxin reductase (FTR) links the regulation of oxygenic photosynthesis to deeply rooted bacteria. Planta 237, 619-635.
DOI URL PMID |
| 7 |
Belin C, Bashandy T, Cela J, Delorme-Hinoux V, Riondet C, Reichheld JP ( 2015). A comprehensive study of thiol reduction gene expression under stress conditions in Ara- bidopsis thaliana.Plant Cell Environ 38, 299-314.
DOI URL PMID |
| 8 |
Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D ( 2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106, 3615-3620.
DOI URL PMID |
| 9 |
Bernal-Bayard P, Ojeda V, Hervás M, Cejudo FJ, Navarro JA, Velázquez-Campoy A, Pérez-Ruiz JM ( 2014). Molecular recognition in the interaction of chloroplast 2-Cys peroxiredoxin with NADPH-thioredoxin reductase C (NTRC) and thioredoxin x. FEBS Lett 588, 4342-4347.
DOI URL PMID |
| 10 |
Betterle N, Ballottari M, Baginsky S, Bassi R ( 2015). High light-dependent phosphorylation of photosystem II inner antenna CP29 in monocots is STN7 independent and enhances nonphotochemical quenching. Plant Physiol 167, 457-471.
DOI URL PMID |
| 11 |
Bohrer AS, Massot V, Innocenti G, Reichheld JP, Issakidis-Bourguet E, Vanacker H ( 2012). New insights into the reduction systems of plastidial thioredoxins point out the unique properties of thioredoxin z from Arabidopsis. J Exp Bot 63, 6315-6323.
DOI URL PMID |
| 12 | Bolter B, Soll J, Schwenkert S ( 2015). Redox meets protein trafficking. Biochim Biophys Acta 1847, 949-956. |
| 13 | Brooks MD, Sylak-Glassman EJ, Fleming GR, Niyogi KK ( 2013). A thioredoxin-like/ β-propeller protein maintains the efficiency of light harvesting in Arabidopsis.Proc Natl Acad Sci USA 110, E2733-E2740. |
| 14 |
Buchanan BB ( 2016). The path to thioredoxin and redox regulation in chloroplasts. Annu Rev Plant Biol 67, 1-24.
DOI URL PMID |
| 15 |
Buchanan BB, Holmgren A, Jacquot JP, Scheibe R ( 2012). Fifty years in the thioredoxin field and a bountiful harvest. Biochim Biophys Acta 1820, 1822-1829.
DOI URL PMID |
| 16 |
Cain P, Hall M, Schröder WP, Kieselbach T, Robinson C ( 2009). A novel extended family of stromal thioredoxins. Plant Mol Biol 70, 273-281.
DOI URL PMID |
| 17 |
Carrillo LR, Froehlich JE, Cruz JA, Savage LJ, Kramer DM ( 2016). Multi-level regulation of the chloroplast ATP synthase: the chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. Plant J 87, 654-663.
DOI URL PMID |
| 18 |
Cejudo FJ, Ferrández J, Cano B, Puerto-Galán L, Guinea M ( 2012). The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signaling. FEBS Lett 586, 2974-2980.
DOI URL PMID |
| 19 |
Chae HB, Moon JC, Shin MR, Chi YH, Jung YJ, Lee SY, Nawkar GM, Jung HS, Hyun JK, Kim WY, Kang CH, Yun DJ, Lee KO, Lee SY ( 2013). Thioredoxin reductase type C (NTRC) orchestrates enhanced thermotolerance to Arabidopsis by its redox-dependent holdase chaperone function. Mol Plant 6, 323-336.
DOI URL PMID |
| 20 |
Cheng F, Zhou YH, Xia XJ, Shi K, Zhou J, Yu JQ ( 2014). Chloroplastic thioredoxin-f and thioredoxin-m1/4 play important roles in brassinosteroids-induced changes in CO2 assimilation and cellular redox homeostasis in tomato. J Exp Bot 65, 4335-4347.
DOI URL PMID |
| 21 |
Chibani K, Tarrago L, Schürmann P, Jacquot JP, Rouhier N ( 2011). Biochemical properties of poplar thioredoxin z. FEBS Lett 585, 1077-1081.
DOI URL PMID |
| 22 |
Cook KM, Hogg PJ ( 2013). Post-translational control of protein function by disulfide bond cleavage. Antioxid Redox Signal 18, 1987-2015.
DOI URL PMID |
| 23 |
Courteille A, Vesa S, Sanz-Barrio R, Cazalé AC, Becuwe- Linka N, Farran I, Havaux M, Rey P, Rumeau D ( 2013). Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol 161, 508-520.
DOI URL PMID |
| 24 |
Couturier J, Chibani K, Jacquot JP, Rouhier N ( 2013). Cysteine-based redox regulation and signaling in plants. Front Plant Sci 4, 105.
DOI URL PMID |
| 25 |
Da QE, Wang P, Wang ML, Sun T, Jin HL, Liu B, Wang JF, Grimm B, Wang HB ( 2017). Thioredoxin and NADPH- dependent thioredoxin reductase C regulation of tetrapyrrole biosynthesis. Plant Physiol 175, 652-666.
DOI URL PMID |
| 26 |
Dangoor I, Peled-Zehavi H, Levitan A, Pasand O, Danon A ( 2009). A small family of chloroplast atypical thioredoxins. Plant Physiol 149, 1240-1250.
DOI URL PMID |
| 27 |
Dangoor I, Peled-Zehavi H, Wittenberg G, Danon A ( 2012). A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24, 1894-1906.
DOI URL PMID |
| 28 | Díaz MG, Hernández-Verdeja T, Kremnev D, Crawford T, Dubreuil C, Strand Å ( 2018). Redox regulation of PEP activity during seedling establishment in Arabidopsis tha- liana.Nat Commun 9, 50. |
| 29 |
Dietz KJ, Pfannschmidt T ( 2011). Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155, 1477-1485.
DOI URL PMID |
| 30 |
Dutton RJ, Wayman A, Wei JR, Rubin EJ, Beckwith J, Boyd D ( 2010). Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci USA 107, 297-301.
DOI URL PMID |
| 31 |
Eliyahu E, Rog I, Inbal D, Danon A ( 2015). ACHT4-driven oxidation of APS1 attenuates starch synthesis under low light intensity in Arabidopsis plants. Proc Natl Acad Sci USA 112, 12876-12881.
DOI URL PMID |
| 32 |
Feng WK, Wang L, Lu Y, Wang XY ( 2011). A protein oxidase catalysing disulfide bond formation is localized to the chloroplast thylakoids. FEBS J 278, 3419-3430.
DOI URL PMID |
| 33 |
Fufezan C, Simionato D, Morosinotto T ( 2012). Identification of key residues for pH dependent activation of violaxanthin de-epoxidase from Arabidopsis thaliana.PLoS One 7, e35669.
DOI URL PMID |
| 34 |
Furt F, Van Oostende C, Widhalm JR, Dale MA, Wertz J, Basset GJC ( 2010). A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K1) in chloroplasts. Plant J 64, 38-46.
DOI URL PMID |
| 35 |
Geigenberger P, Fernie AR ( 2014). Metabolic control of redox and redox control of metabolism in plants. Antioxid Redox Signal 21, 1389-1421.
DOI URL PMID |
| 36 | Goss R, Lepetit B ( 2015). Biodiversity of NPQ. J Plant Physiol 172, 13-32. |
| 37 |
Gütle DD, Roret T, Hecker A, Reski R, Jacquot JP ( 2017). Dithiol disulphide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci 255, 1-11.
DOI URL PMID |
| 38 |
Gütle DD, Roret T, Müller SJ, Couturier J, Lemaire SD, Hecker A, Dhalleine T, Buchanan BB, Reski R, Einsle O, Jacquot JP ( 2016). Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc Natl Acad Sci USA 113, 6779-6784.
DOI URL PMID |
| 39 |
Hall M, Mata-Cabana A, Åkerlund HE, Florencio FJ, Schröder WP, Lindahl M, Kieselbach T ( 2010). Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10, 987-1001.
DOI URL PMID |
| 40 |
Hallin EI, Guo K, Åkerlund HE ( 2015). Violaxanthin de-epo- xidase disulphides and their role in activity and thermal stability. Photosynth Res 124, 191-198.
DOI URL PMID |
| 41 |
Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D ( 2013). PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49, 511-523.
DOI URL PMID |
| 42 | Hisabori T, Sunamura EI, Kim Y, Konno H ( 2013). The chloroplast ATP synthase features the characteristic redox regulation machinery. Antioxid Redox Signal 19, 1846-1854. |
| 43 |
Jacquot JP, Eklund H, Rouhier N, Schürmann P ( 2009). Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci 14, 336-343.
DOI URL |
| 44 |
Järvi S, Gollan PJ, Aro EM ( 2013). Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front Plant Sci 4, 434.
DOI URL PMID |
| 45 |
Kang ZH, Wang GX ( 2016). Redox regulation in the thylakoid lumen. J Plant Physiol 192, 28-37.
DOI URL PMID |
| 46 |
Karamoko M, Cline S, Redding K, Ruiz N, Hamel PP ( 2011). Lumen Thiol Oxidoreductase 1, a disulfide bond- forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23, 4462-4475.
DOI URL PMID |
| 47 | Karamoko M, Gabilly ST, Hamel PP ( 2013). Operation of trans-thylakoid thiol-metabolizing pathways in photosynthesis.Front Plant Sci 4, 476. |
| 48 | Kieselbach T ( 2013). Oxidative folding in chloroplasts. Antioxid Redox Signal 19, 72-82. |
| 49 | Kim MR, Khaleda L, Jung IJ, Kim JY, Lee SY, Cha JY, Kim WY ( 2017). Overexpression of chloroplast-localized NAD- PH-dependent thioredoxin reductase C (NTRC) enhances tolerance to photo-oxidative and drought stresses in Ara- bidopsis thaliana.J Plant Biol 60, 175-180. |
| 50 | Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ ( 2012). NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell 24, 1534-1548. |
| 51 | Kirchsteiger K, Pulido P, González M, Cejudo FJ ( 2009). NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thalia- na.Mol Plant 2, 298-307. |
| 52 |
König J, Muthuramalingam M, Dietz KJ ( 2012). Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr Opin Plant Biol 15, 261-268.
DOI URL PMID |
| 53 |
Laugier E, Tarrago L, Courteille A, Innocenti G, Eymery F, Rumeau D, Issakidis-Bourguet E, Rey P ( 2013). Involvement of thioredoxin y2 in the preservation of leaf methionine sulfoxide reductase capacity and growth under high light. Plant Cell Environ 36, 670-682.
DOI URL PMID |
| 54 |
Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E ( 2009). Chlo- roplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149, 1261-1276.
DOI URL PMID |
| 55 |
Lindahl M, Kieselbach T ( 2009). Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J Proteomics 72, 416-438.
DOI URL PMID |
| 56 |
Lindahl M, Mata-Cabana A, Kieselbach T ( 2011). The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal 14, 2581-2642.
DOI URL PMID |
| 57 |
Lu Y, Wang HR, Li H, Cui HR, Feng YG, Wang XY ( 2013). A chloroplast membrane protein LTO1/AtVKOR involving in redox regulation and ROS homeostasis. Plant Cell Rep 32, 1427-1440.
DOI URL PMID |
| 58 | Luo T, Fan TT, Liu YN, Rothbart M, Yu J, Zhou SX, Grimm B, Luo MZ ( 2012). Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol 159, 118-130. |
| 59 |
Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C ( 2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17, 1124-1160.
DOI URL PMID |
| 60 | Meyer Y, Buchanan BB, Vignols F, Reichheld JP ( 2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43, 335-367. |
| 61 |
Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P ( 2009). NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106, 9908-9913.
DOI URL |
| 62 |
Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez- Pérez ME, Francia F, Danon A, Marchand CH, Fermani S, Trost P, Lemaire SD ( 2013). Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 4, 470.
DOI URL PMID |
| 63 |
Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB ( 2009). Thioredoxin targets in plants: the first 30 years. J Proteomics 72, 452-474.
DOI URL PMID |
| 64 |
Motohashi K, Hisabori T ( 2010). CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid Redox Signal 13, 1169-1176.
DOI URL PMID |
| 65 |
Naranjo B, Díaz-Espejo A, Lindahl M, Cejudo FJ ( 2016 a). Type- f thioredoxins have a role in the short-term activation of carbon metabolism and their loss affects growth under short-day conditions in Arabidopsis thaliana.J Exp Bot 67, 1951-1964.
DOI URL PMID |
| 66 |
Naranjo B, Mignée C, Krieger-Liszkay A, Hornero-Méndez D, Gallardo-Guerrero L, Cejudo FJ, Lindahl M ( 2016 b). The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ 39, 804-822.
DOI URL PMID |
| 67 |
Née G, Zaffagnini M, Trost P, Issakidis-Bourguet E ( 2009). Redox regulation of chloroplastic glucose-6-phosphate dehydrogenase: a new role for f-type thioredoxin. FEBS Lett 583, 2827-2832.
DOI URL PMID |
| 68 |
Nikkanen L, Rintamäki E ( 2014). Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos Trans Roy Soc Lond B Biol Sci 369, 20130224.
DOI URL PMID |
| 69 |
Nikkanen L, Toivola J, Diaz MG, Rintamäki E ( 2017). Chloroplast thioredoxin systems: prospects for improving photosynthesis. Philos Trans Roy Soc Lond B Biol Sci 372, 20160474.
DOI URL PMID |
| 70 |
Nikkanen L, Toivola J, Rintamäki E ( 2016). Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant Cell Environ 39, 1691-1705.
DOI URL PMID |
| 71 |
Ojeda V, Pérez-Ruiz JM, González M, Nájera VA, Sahrawy M, Serrato AJ, Geigenberger P, Cejudo FJ ( 2017). NADPH thioredoxin reductase C and thioredoxins act concertedly in seedling development. Plant Physiol 174, 1436-1448.
DOI URL PMID |
| 72 |
Okegawa Y, Motohashi K ( 2015). Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo.Plant J 84, 900-913.
DOI URL PMID |
| 73 |
Onoa B, Schneider AR, Brooks MD, Grob P, Nogales E, Geissler PL, Niyogi KK, Bustamante C ( 2014). Atomic force microscopy of photosystem II and its unit cell clustering quantitatively delineate the mesoscale variability in Arabidopsis thylakoids. PLoS One 9, e101470.
DOI URL PMID |
| 74 |
Pérez-Ruiz JM, González M, Spínola MC, Sandalio LM, Cejudo FJ ( 2009). The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Mol Plant 2, 457-467.
DOI URL PMID |
| 75 |
Pérez-Ruiz JM, Guinea M, Puerto-Galán L, Cejudo FJ ( 2014). NADPH thioredoxin reductase C is involved in redox regulation of the Mg-chelatase I subunit in Arabidopsis thaliana chloroplasts.Mol Plant 7, 1252-1255.
DOI URL PMID |
| 76 |
Pérez-Ruiz JM, Naranjo B, Ojeda V, Guinea M, Cejudo FJ ( 2017). NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc Natl Acad Sci USA 114, 12069-12074.
DOI URL PMID |
| 77 |
Puerto-Galán L, Pérez-Ruiz JM, Guinea M, Cejudo FJ ( 2015). The contribution of NADPH thioredoxin reductase C (NTRC) and sulfiredoxin to 2-Cys peroxiredoxin overoxidation in Arabidopsis thaliana chloroplasts.J Exp Bot 66, 2957-2966.
DOI URL PMID |
| 78 |
Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ ( 2010). Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts.J Exp Bot 61, 4043-4054.
DOI URL PMID |
| 79 |
Rey P, Sanz-Barrio R, Innocenti G, Ksas B, Courteille A, Rumeau D, Issakidis-Bourguet E, Farran I ( 2013). Overexpression of plastidial thioredoxins f and m differentially alters photosynthetic activity and response to oxidative stress in tobacco plants. Front Plant Sci 4, 390.
DOI URL PMID |
| 80 |
Richter AS, Grimm B ( 2013). Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway in plants. Front Plant Sci 4, 371.
DOI URL PMID |
| 81 | Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamäki E, Grimm B ( 2013). Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol 162, 63-73. |
| 82 |
Rouhier N, Cerveau D, Couturier J, Reichheld JP, Rey P ( 2015). Involvement of thiol-based mechanisms in plant development. Biochim Biophys Acta 1850, 1479-1496.
DOI URL PMID |
| 83 |
Sanz-Barrio R, Corral-Martinez P, Ancin M, Segui- Simarro JM, Farran I ( 2013). Overexpression of plastidial thioredoxin f leads to enhanced starch accumulation in tobacco leaves. Plant Biotechnol J 11, 618-627.
DOI URL PMID |
| 84 |
Sanz-Barrio R, Fernández-San Millán A, Carballeda J, Corral-Martínez P, Seguí-Simarro JM, Farran I ( 2012). Chaperone-like properties of tobacco plastid thioredoxins f and m. J Exp Bot 63, 365-379.
DOI URL PMID |
| 85 |
Serrato AJ, Fernández-Trijueque J, Barajas-López JDD, Chueca A, Sahrawy M ( 2013). Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front Plant Sci 4, 463.
DOI URL PMID |
| 86 |
Simionato D, Basso S, Zaffagnini M, Lana T, Marzotto F, Trost P, Morosinotto T ( 2015). Protein redox regulation in the thylakoid lumen: the importance of disulfide bonds for violaxanthin de-epoxidase. FEBS Lett 589, 919-923.
DOI URL PMID |
| 87 |
Stenbaek A, Jensen PE ( 2010). Redox regulation of chlorophyll biosynthesis. Phytochemistry 71, 853-859.
DOI URL PMID |
| 88 |
Strand DD, Fisher N, Davis GA, Kramer DM ( 2016). Redox regulation of the antimycin A sensitive pathway of cyclic electron flow around photosystem I in higher plant thylakoids. Biochim Biophys Acta 1857, 1-6.
DOI URL PMID |
| 89 |
Tarrago L, Laugier E, Zaffagnini M, Marchand CH, Le Maréchal P, Lemaire SD, Rey P ( 2010). Plant thioredoxin CDSP32 regenerates 1-cys methionine sulfoxide reductase B activity through the direct reduction of sulfenic acid. J Biol Chem 285, 14964-14972.
DOI URL PMID |
| 90 | Thormählen I, Meitzel T, Groysman J, Öchsner AB, von Roepenack-Lahaye E, Naranjo B, Cejudo FJ, Geigenberger P ( 2015). Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiol 169, 1766-1786. |
| 91 |
Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis- Bourguet E, Geigenberger P ( 2013). Inactivation of thioredoxin f1 leads to decreased light activation of ADP- glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36, 16-29.
DOI URL PMID |
| 92 |
Thormählen I, Zupok A, Rescher J, Leger J, Weissenberger S, Groysman J, Orwat A, Chatel-Innocenti G, Issakidis-Bourguet E, Armbruster U, Geigenberger P ( 2017). Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol Plant 10, 168-182.
DOI URL |
| 93 |
Tikkanen M, Grieco M, Kangasjärvi S, Aro EM ( 2010). Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol 152, 723-735.
DOI URL |
| 94 |
Toivola J, Nikkanen L, Dahlström KM, Salminen TA, Lepistö A, Vignols F, Rintamäki E ( 2013). Overexpression of chloroplast NADPH-dependent thioredoxin reductase in Arabidopsis enhances leaf growth and elucidates in vivo function of reductase and thioredoxin domains.Front Plant Sci 4, 389.
DOI URL PMID |
| 95 |
Wang P, Liu J, Liu B, Da QE, Feng DR, Su JB, Zhang Y, Wang JF, Wang HB ( 2014). Ferredoxin: thioredoxin reductase is required for proper chloroplast development and is involved in the regulation of plastid gene expression in Arabidopsis thaliana.Mol Plant 7, 1586-1590.
DOI URL PMID |
| 96 |
Wang P, Liu J, Liu B, Feng DR, Da QE, Wang P, Shu SY, Su JB, Zhang Y, Wang Jf, Wang HB ( 2013). Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol 163, 1710-1728.
DOI URL PMID |
| 97 |
Wang XY, Dutton RJ, Beckwith J, Boyd D ( 2011). Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase.Antioxid Redox Signal 14, 1413-1420.
DOI URL PMID |
| 98 |
Wulff RP, Lundqvist J, Rutsdottir G, Hansson A, Stenbaek A, Elmlund D, Elmlund H, Jensen PE, Hansson M ( 2011). The activity of barley NADPH-dependent thioredoxin reductase C is independent of the oligomeric state of the protein: tetrameric structure determined by cryoelec- tron microscopy. Biochemistry 50, 3713-3723.
DOI URL PMID |
| 99 | Yoshida K, Hara S, Hisabori T ( 2015). Thioredoxin selecti- vity for thiol-based redox regulation of target proteins in chloroplasts. J Biol Chem 290, 14278-14288. |
| 100 |
Yoshida K, Hisabori T ( 2016). Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc Natl Acad Sci USA 113, E3967-E3976.
DOI URL PMID |
| 101 |
Yoshida K, Hisabori T ( 2017). Distinct electron transfer from ferredoxin-thioredoxin reductase to multiple thioredoxin isoforms in chloroplasts. Biochem J 474, 1347-1360.
DOI URL |
| 102 |
Yoshida K, Matsuoka Y, Hara S, Konno H, Hisabori T ( 2014). Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana.Plant Cell Physiol 55, 1415-1425.
DOI URL PMID |
| [1] | Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome and Phylogenetic Inference of the Aronia melanocarpa [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | YIN Si, YANG Yi-Ting, LU Rui-Ling, NIAN Rui, HAO Zhuan, GAO Yong. Phylogeographic study of natural populations of Amorphophallus yunnanensis (Araceae) in China [J]. Chin J Plant Ecol, 2025, 49(2): 308-319. |
| [3] | Xiaolin Yu, Xiya Li, Bingyujie Xia, Hao Li, Baocai Tan, Yong Wang. Research Advance of PPR Proteins Involved in the Mechanism of Organelle RNA C→U Editing [J]. Chinese Bulletin of Botany, 2024, 59(6): 903-911. |
| [4] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [5] | Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian. Comparative Analyses on the Chloroplast Genome of Three Sympatric Atraphaxis Species [J]. Chinese Bulletin of Botany, 2023, 58(3): 417-432. |
| [6] | Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li, Qianqian Deng, Yingzi Li, Xinmin Tian. Analysis of Chloroplast Genomes of Aleurites moluccana [J]. Chinese Bulletin of Botany, 2023, 58(2): 248-260. |
| [7] | Qi Zhang, Wenjing Zhang, Xiankai Yuan, Ming Li, Qiang Zhao, Yanli Du, Jidao Du. The Regulatory Mechanism of Melatonin on Nucleic Acid Repairing of Common Bean (Phaseolus vulgaris) at the Sprout Stage Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 108-121. |
| [8] | Li Chen, Liu Jianting, Fan Yongxin, Zhao Xuehui, Xiao Wei, Chen Xiude, Fu Xiling, Li Ling, Li Dongmei. Effects of UV-B on Photosynthetic Function and Chloroplast Ultrastructure of Peach Leaves Grown in Greenhouse [J]. Chinese Bulletin of Botany, 2022, 57(4): 434-443. |
| [9] | Kairu Yang, Qiwei Jia, Jiayi Jin, Hanfei Ye, Sheng Wang, Qianyu Chen, Yian Guan, Chenyang Pan, Dedong Xin, Yuan Fang, Yuexing Wang, Yuchun Rao. Cloning and Functional Analysis of Rice Yellow Green Leaf Regulatory Gene YGL18 [J]. Chinese Bulletin of Botany, 2022, 57(3): 276-287. |
| [10] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| [11] | Menghua Zhang, Xianchun Zhang. Species delimitation of the Selaginella delicatula group in China [J]. Biodiv Sci, 2021, 29(12): 1607-1619. |
| [12] | Yan Wang, Bowei Jia, Mingzhe Sun, Xiaoli Sun. Advances in Molecular Mechanisms of Stress Tolerance in Wild Soybean [J]. Chinese Bulletin of Botany, 2021, 56(1): 104-115. |
| [13] | WANG Chun-Cheng, MA Song-Mei, ZHANG Dan, WANG Shao-Ming. Spatial genetic structure of Lycium ruthenicum in the Qaidam Basin [J]. Chin J Plant Ecol, 2020, 44(6): 661-668. |
| [14] | Chunyan Zhang. The Measurement Methods and Principles of P700 Redox Kinetics [J]. Chinese Bulletin of Botany, 2020, 55(6): 740-748. |
| [15] | Yuemei Zhao,Zhenyan Yang,Yongping Zhao,Xiaoling Li,Zhixin Zhao,Guifang Zhao. Chloroplast Genome Structural Characteristics and Phylogenetic Relationships of Oleaceae [J]. Chinese Bulletin of Botany, 2019, 54(4): 441-454. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||