

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (3): 276-287.DOI: 10.11983/CBB22018 cstr: 32102.14.CBB22018
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Kairu Yang1, Qiwei Jia1, Jiayi Jin1, Hanfei Ye1, Sheng Wang1, Qianyu Chen1, Yian Guan1, Chenyang Pan1, Dedong Xin1, Yuan Fang1,*( ), Yuexing Wang2,*(
), Yuexing Wang2,*( ), Yuchun Rao1,*(
), Yuchun Rao1,*( )
)
Received:2022-01-19
Accepted:2022-03-18
Online:2022-05-01
Published:2022-05-18
Contact:
Yuan Fang,Yuexing Wang,Yuchun Rao
Kairu Yang, Qiwei Jia, Jiayi Jin, Hanfei Ye, Sheng Wang, Qianyu Chen, Yian Guan, Chenyang Pan, Dedong Xin, Yuan Fang, Yuexing Wang, Yuchun Rao. Cloning and Functional Analysis of Rice Yellow Green Leaf Regulatory Gene YGL18[J]. Chinese Bulletin of Botany, 2022, 57(3): 276-287.
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| B3-14 | AGGATTTGGGCTGTCAATGC | AGCCTACCACAAGACATATAATCG |
| B3-15 | TCCAGTCCACTAAAAGTTTTC | ATTATGCTATAGGCTTTGAGC |
| M1 | TGGAAATGGGTGGAATACCT | CCTCTGTGAGGACCATCTCC |
| M2 | GAATTATTTTTCCTTGGTCATTGTG | GTACCAACGAGGGTCCAAAG |
| M3 | GGAGGCAGGCTACACCAGTA | GCACAATTCTTAAAGAGATGAACC |
| M4 | TTTGATTGGGCTCAAAGCAC | GCCAGGATTTGTTGGATTAGG |
| M5 | AAAAACTCCCAGCGGATAGA | GCTAGAACTGGCAAAATGCTG |
| Indel1 | TTAGGCAAATGAAACCAAGG | CCCAACCACCTAAGTAAACGA |
| Indel2 | TCCTTTGATGACTGGTCGGC | ATGGGAAGACGGGGGAGTAA |
| Indel3 | GTTTGGGAGAGACATGGGGG | GTTTGGCCCATTCCCTGAAG |
| Indel4 | ATGATTTGGAGCCAGCAGTT | GGAAGTGGAATTGCAGATGG |
Table 1 Primer sequences for fine mapping
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| B3-14 | AGGATTTGGGCTGTCAATGC | AGCCTACCACAAGACATATAATCG |
| B3-15 | TCCAGTCCACTAAAAGTTTTC | ATTATGCTATAGGCTTTGAGC |
| M1 | TGGAAATGGGTGGAATACCT | CCTCTGTGAGGACCATCTCC |
| M2 | GAATTATTTTTCCTTGGTCATTGTG | GTACCAACGAGGGTCCAAAG |
| M3 | GGAGGCAGGCTACACCAGTA | GCACAATTCTTAAAGAGATGAACC |
| M4 | TTTGATTGGGCTCAAAGCAC | GCCAGGATTTGTTGGATTAGG |
| M5 | AAAAACTCCCAGCGGATAGA | GCTAGAACTGGCAAAATGCTG |
| Indel1 | TTAGGCAAATGAAACCAAGG | CCCAACCACCTAAGTAAACGA |
| Indel2 | TCCTTTGATGACTGGTCGGC | ATGGGAAGACGGGGGAGTAA |
| Indel3 | GTTTGGGAGAGACATGGGGG | GTTTGGCCCATTCCCTGAAG |
| Indel4 | ATGATTTGGAGCCAGCAGTT | GGAAGTGGAATTGCAGATGG |
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CAO1-qrt | GCTGCTCTACCGGATGTCTC | ACAACCGATACCGGATACCA |
| DVR-qrt | TTCACCTACAGCATCGTCCG | AACAGCATCTCCCCTTGCTC |
| YGL1-qrt | TGTTGGTGGGTCCTTGCTTT | ACTGAAGCCCCAGAGCTCTA |
| PORA-qrt | CATGCTCGACGACCTCAAGA | TCCTGCATCGTCAGCATGTT |
| PORB-qrt | CATCATCGTCGGCTCCATCA | TCCTGCATCGTCAGCATGTT |
| ChlH-qrt | GGGAACTTGGCGTTTCATTA | TCAAATGTCTTGCGTTGCTC |
| ChlD-qrt | TCCTTTAGCTCACGGCCTTA | GTCCAACATCACCGCTCTTT |
| NOL-qrt | TCACAAGCCTTACGACCCAC | GCTCCCTCTCCATCATCTGC |
| NYC1-qrt | TAGTTGGCTCGGGGGAGTAA | AAGGACTGATTCAGGGCTGC |
| OsFdC2-qrt | AGCTGTGCGGTTCGGATAAA | AACAGGTCCTCGTGCGAAAT |
| rpoA-qrt | CCATTCCCACAAGCAAAAAT | TCTTACCGCCTTCCGTAGAA |
| rpoB-qrt | TGGTACATATCCCTTATCTCAA | CTCCAGGACCCAAACAACTC |
| rbcL-qrt | CTTGGCAGCATTCCGAGTAA | ACAACGGGCTCGATGTGATA |
| rbcS-qrt | GCTTGGAGTTCAGCAAGGTC | AACGAAGGCATCAGGGTATG |
| PsaA-qrt | TTAGAAATCCGCCAATCCA | TGCTAGGCTCTACAACCATT |
| PsaD-qrt | CCGCTCCAAGTACAAGATCA | AAGAGCAGCCTGACAGATGA |
| PsbA-qrt | ACCCTCATTAGCAGATTCGT | GATTGTATTCCAGGCAGAGC |
| PsbD-qrt | AAGACAGATTCCGAGGGTGG | TGATTCGCTAGGGATTAAAGAG |
Table 2 Primer sequences for qRT-PCR
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CAO1-qrt | GCTGCTCTACCGGATGTCTC | ACAACCGATACCGGATACCA |
| DVR-qrt | TTCACCTACAGCATCGTCCG | AACAGCATCTCCCCTTGCTC |
| YGL1-qrt | TGTTGGTGGGTCCTTGCTTT | ACTGAAGCCCCAGAGCTCTA |
| PORA-qrt | CATGCTCGACGACCTCAAGA | TCCTGCATCGTCAGCATGTT |
| PORB-qrt | CATCATCGTCGGCTCCATCA | TCCTGCATCGTCAGCATGTT |
| ChlH-qrt | GGGAACTTGGCGTTTCATTA | TCAAATGTCTTGCGTTGCTC |
| ChlD-qrt | TCCTTTAGCTCACGGCCTTA | GTCCAACATCACCGCTCTTT |
| NOL-qrt | TCACAAGCCTTACGACCCAC | GCTCCCTCTCCATCATCTGC |
| NYC1-qrt | TAGTTGGCTCGGGGGAGTAA | AAGGACTGATTCAGGGCTGC |
| OsFdC2-qrt | AGCTGTGCGGTTCGGATAAA | AACAGGTCCTCGTGCGAAAT |
| rpoA-qrt | CCATTCCCACAAGCAAAAAT | TCTTACCGCCTTCCGTAGAA |
| rpoB-qrt | TGGTACATATCCCTTATCTCAA | CTCCAGGACCCAAACAACTC |
| rbcL-qrt | CTTGGCAGCATTCCGAGTAA | ACAACGGGCTCGATGTGATA |
| rbcS-qrt | GCTTGGAGTTCAGCAAGGTC | AACGAAGGCATCAGGGTATG |
| PsaA-qrt | TTAGAAATCCGCCAATCCA | TGCTAGGCTCTACAACCATT |
| PsaD-qrt | CCGCTCCAAGTACAAGATCA | AAGAGCAGCCTGACAGATGA |
| PsbA-qrt | ACCCTCATTAGCAGATTCGT | GATTGTATTCCAGGCAGAGC |
| PsbD-qrt | AAGACAGATTCCGAGGGTGG | TGATTCGCTAGGGATTAAAGAG |
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CRISPR/Cas9 | CTCCCTCTCTCGATCGTCAG | GCTACTGCTGCTTGAGACGG |
Table 3 Primer sequences for CRISPR/Cas9
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CRISPR/Cas9 | CTCCCTCTCTCGATCGTCAG | GCTACTGCTGCTTGAGACGG |

Figure 1 Phenotypic of wild-type HZ and mutant ygl18 of rice (A) Phenotype of plant at tillering stage (bar=6 cm); (B) Phenotype of plant at maturity stage (bar=10 cm)
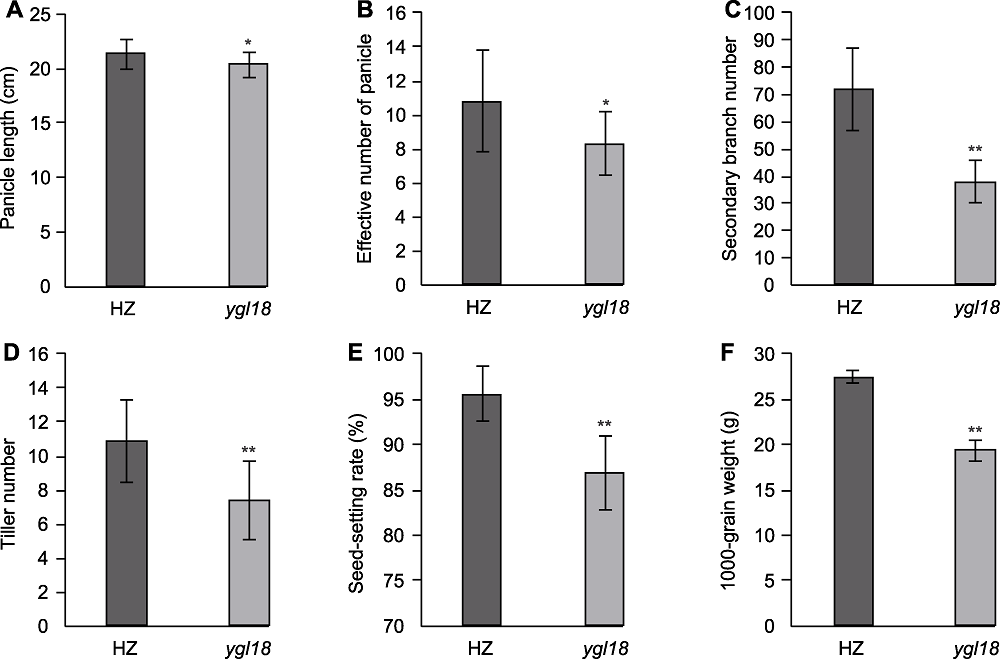
Figure 2 Agronomic characters of wild-type HZ and mutant ygl18 of rice (A) Panicle length; (B) Effective number of panicle; (C) Secondary branch number; (D) Tiller number; (E) Seed-setting rate; (F) 1000-grain weight. * and ** indicate significant differences at 0.05 and 0.01 levels, respectively.
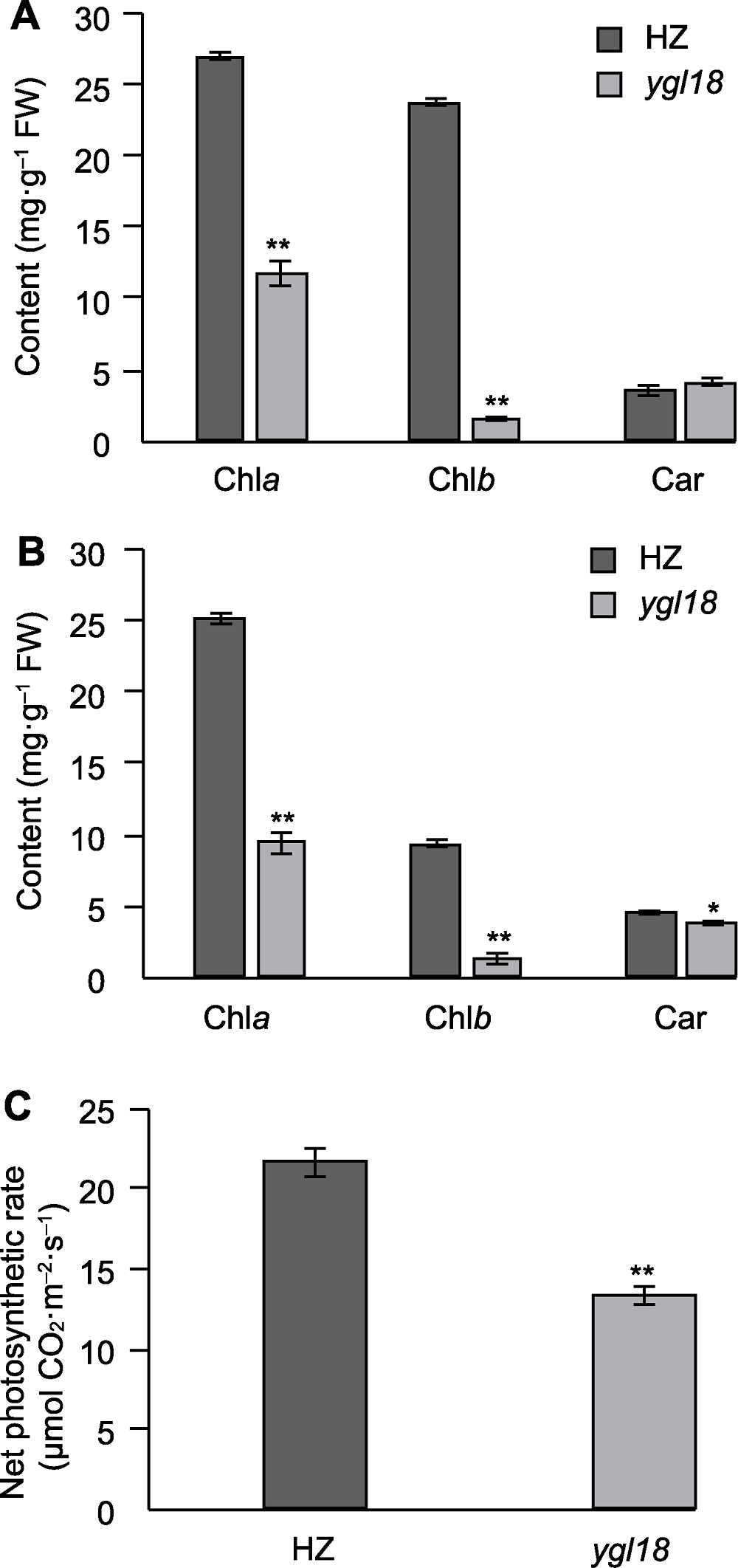
Figure 3 Photosynthetic pigment contents and net photosynthetic rate in leaves of wild-type HZ and mutant ygl18 of rice (A) Photosynthetic pigment content of leaves at tillering stage; (B) Photosynthetic pigment content of leaves at heading stage; (C) Net photosynthetic rate of leaves. * and ** indicate significant differences at 0.05 and 0.01 levels, respectively.
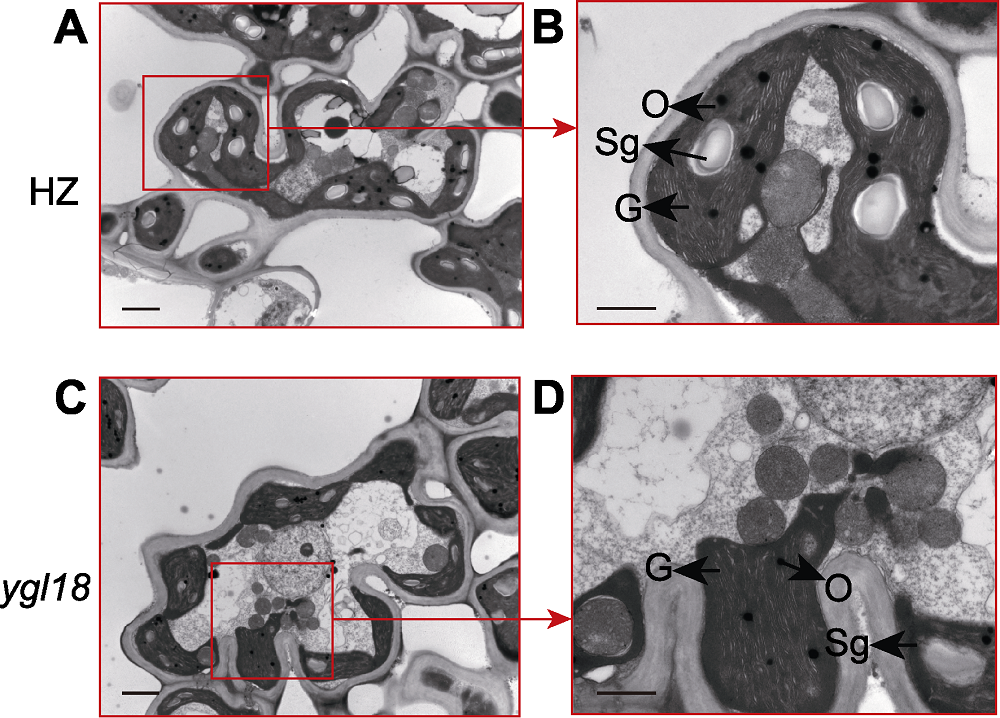
Figure 4 Ultrastructural observation of mesophyll cells in wild-type HZ and mutant ygl18 of rice (A) HZ mesophyll cells (bar=2 μm); (B) HZ mesophyll cells (bar=1 μm); (C) ygl18 mesophyll cells (bar=2 μm); (D) ygl18 mesophyll cells (bar=1 μm). G: Grana lamella; Sg: Starch granule; O: Ophilic granule
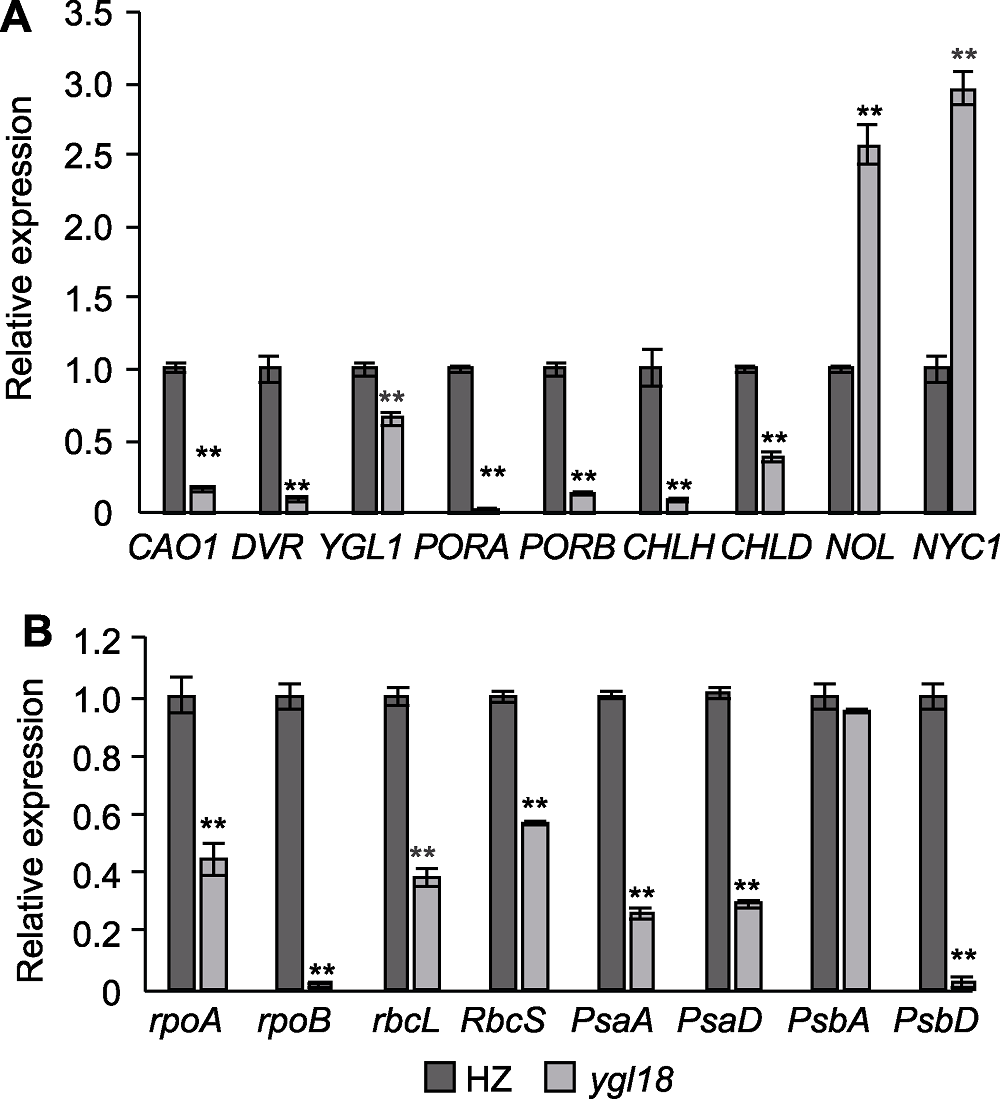
Figure 5 Expression levels of chlorophyll synthesis, chloroplast development and photosynthesis related genes in wild-type HZ and mutant ygl18 of rice at tillering stage (A) Expression of genes related to chlorophyll synthesis; (B) Expression of genes related to chloroplast development and photosynthesis
| Cross | No. of F2 individuals | χ20.05<3.841 | ||
|---|---|---|---|---|
| Normal | yellow-green leaf | Total | ||
| ygl18 × NIP | 836 | 331 | 1167 | 0.833 |
Table 4 Genetic analysis of F2 population of rice mutant
| Cross | No. of F2 individuals | χ20.05<3.841 | ||
|---|---|---|---|---|
| Normal | yellow-green leaf | Total | ||
| ygl18 × NIP | 836 | 331 | 1167 | 0.833 |
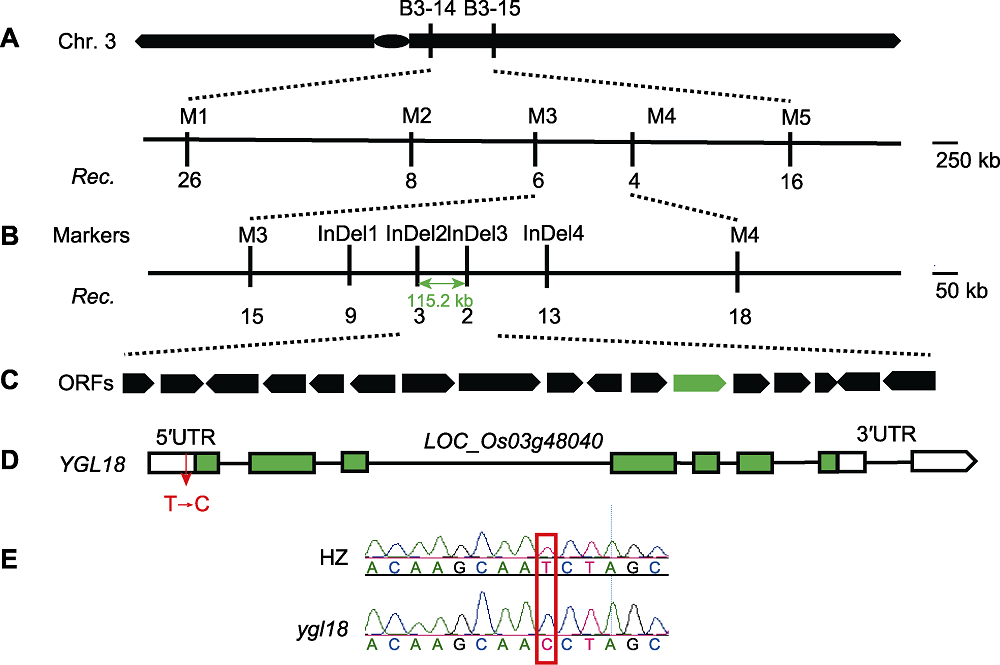
Figure 6 Fine mapping of YGL18 in rice (A) YGL18 was located between B3-14 and B3-15 on long arm of rice chromosome 3; (B) Then it was narrowed to a region between M3 and M4 using 148 F2 mutants; (C) Then it was limited to an area of about 115.2 kb between InDel2 and InDel3 with a total of 15 open reading frames (ORFs), using 504 F2 mutants; (D) YGL18 gene structure modeling (green boxes indicate exons, white boxes indicate UTRs, and black lines between boxes indicate introns); (E) Peak sequencing result of YGL18 gene between wild type and mutant
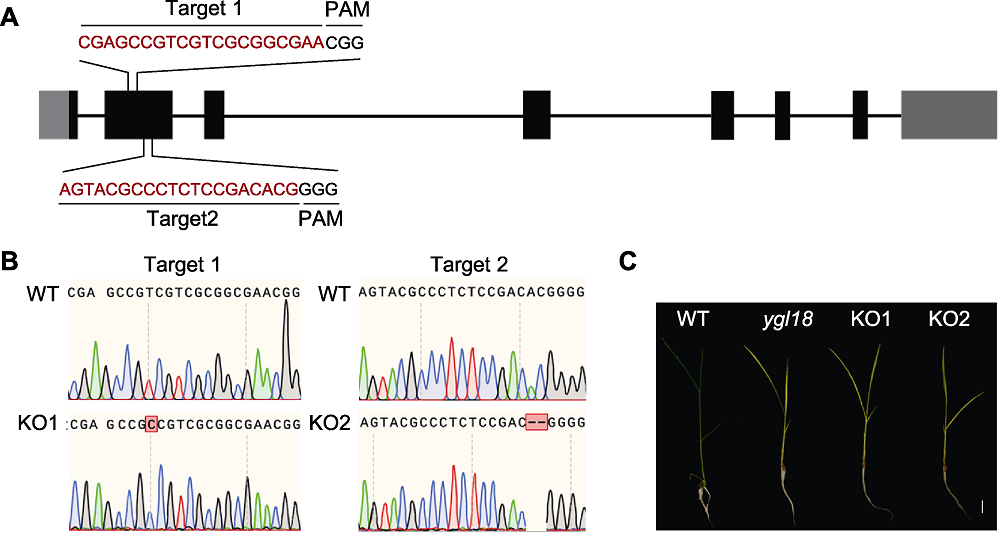
Figure 7 CRISPR/Cas9 knockout verification of YGL18 in rice (A) The position of two separate knockout targets (black boxes represent exons, gray boxes represent UTRs, black lines represent introns, and Target 1 and Target 2 are two separate targets); (B) Sequencing verification of gene knockout strains; (C) Phenotype of wild type (WT), mutant ygl18, KO1 and KO2 lines (bar=2 cm). PAM: Protospacer adjacent motif
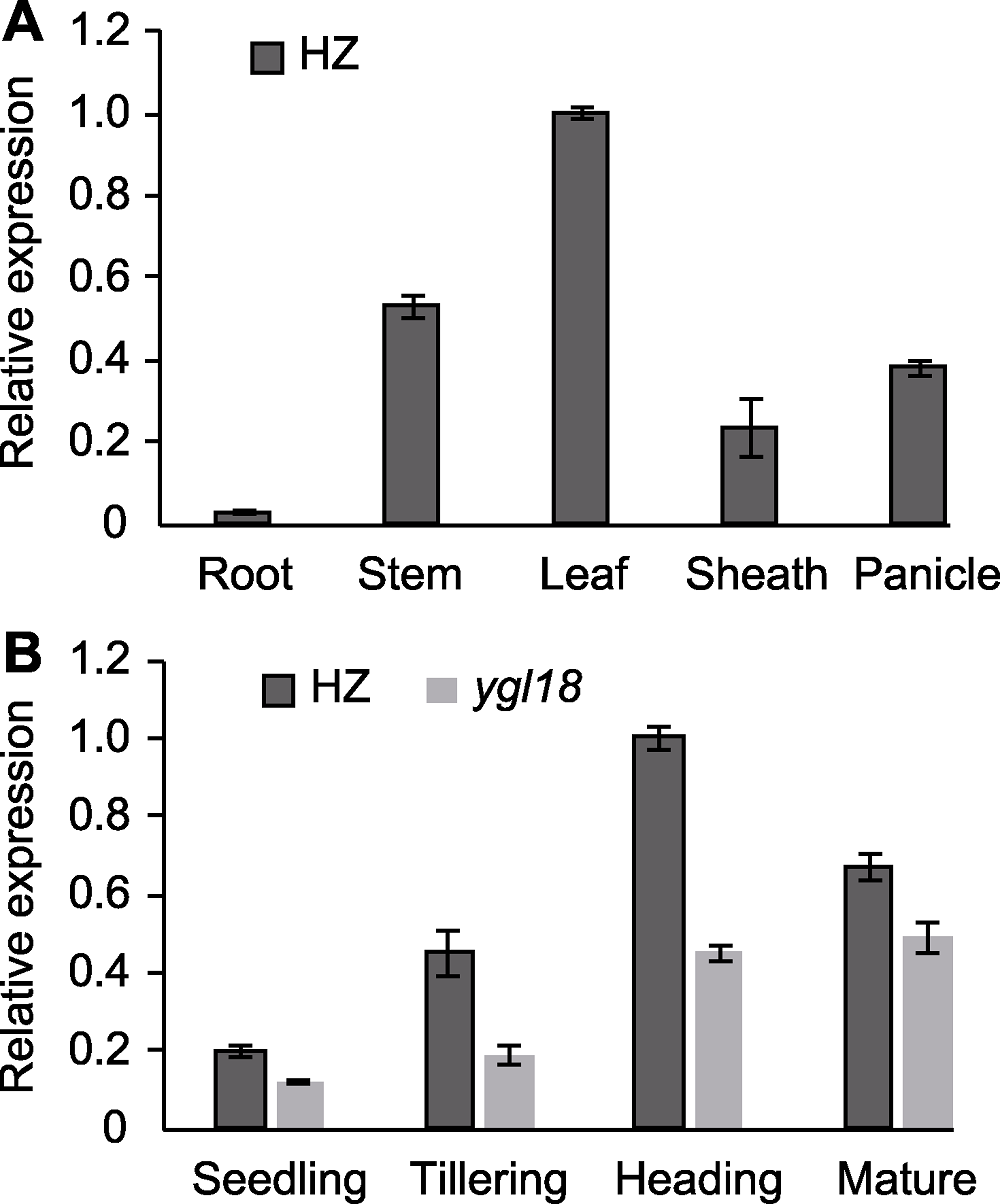
Figure 8 Expression pattern of gene YGL18 in wild-type HZ and mutant ygl18 of rice (A) Tissue specific expression of YGL18; (B) The expression level of YGL18 in different development stage
| [1] | 陈昆松, 李方, 徐昌杰, 张上隆, 傅承新 (2004). 改良CTAB法用于多年生植物组织基因组DNA的大量提取. 遗传 26, 529-531. |
| [2] | 杜芳芳, 马骏杰, 杨泽伟, 赵雪莲, 刘东宇, 杨秀芹 (2021). 5'UTR在基因表达调控中的研究进展. 中国畜牧杂志 57(8), 60-67. |
| [3] | 赫磊 (2020). 铁氧还蛋白调控水稻光合电子传递的分子机理研究. 博士论文. 北京: 中国农业科学院. pp. 1-101. |
| [4] |
林添资, 孙立亭, 景德道, 钱华飞, 余波, 曾生元, 李闯, 龚红兵 (2018). 一个水稻黄绿叶突变体ygl14(t)的鉴定及基因定位. 核农学报 32, 216-226.
DOI |
| [5] | 任永梅, 王嘉宇, 王琬璐, 王棋, 姜鑫, 刘振宇, 朱琳 (2019). 水稻白条纹叶突变体wsl1的生理特性分析及基因定位. 沈阳农业大学学报 50, 87-92. |
| [6] | 韦敏益, 吴子帅, 刘立龙, 李允振, 农春晓, 覃宝祥, 李容柏 (2016). 一个水稻长芒基因AWN-2的定位与克隆. 基因组学与应用生物学 35, 949-956. |
| [7] |
徐娜, 徐江民, 蒋玲欢, 饶玉春 (2017). 水稻叶片早衰成因及分子机理研究进展. 植物学报 52, 102-112.
DOI |
| [8] | 许子怡, 程行, 沈奇, 赵亚男, 汤佳玉, 刘喜 (2021). 水稻黄绿叶突变体ygl3的鉴定与基因功能分析. 中国农业科学 54, 3149-3157. |
| [9] | 杨颜榕, 黄纤纤, 赵亚男, 汤佳玉, 刘喜 (2020). 水稻叶色基因克隆与分子机制研究进展. 植物遗传资源学报 21, 794- 803. |
| [10] | 张芳燕, 罗翔, 董娜, 张鹏 (2011). 5'UTR与基因表达的关系. 科技经济市场 (3), 13-16. |
| [11] | 张力科, 高用明 (2009). 水稻叶色突变体及其基因定位和克隆的研究进展. 作物杂志 (2), 12-16. |
| [12] |
周纯, 焦然, 胡萍, 林晗, 胡娟, 徐娜, 吴先美, 饶玉春, 王跃星 (2019). 水稻早衰突变体LS-es1的基因定位及候选基因分析. 植物学报 54, 606-619.
DOI |
| [13] |
周亭亭, 饶玉春, 任德勇 (2018). 水稻卷叶细胞学与分子机制研究进展. 植物学报 53, 848-855.
DOI |
| [14] | Akhter D (2019). 水稻叶色基因OsBML和OsPL的基因定位和功能分析. 博士论文. 杭州: 浙江大学. pp. 52-54. |
| [15] |
Chiu FY, Chen YR, Tu SL (2010). Electrostatic interaction of phytochromobilin synthase and ferredoxin for biosynthesis of phytochrome chromophore. J Biol Chem 285, 5056-5065.
DOI URL |
| [16] |
Dong H, Fei GL, Wu CY, Wu FQ, Sun YY, Chen MJ, Ren YL, Zhou KN, Cheng ZJ, Wang JL, Jiang L, Zhang X, Guo XP, Lei CL, Su N, Wang HY, Wan JM (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol 162, 1867-1880.
DOI PMID |
| [17] |
Gou P, Hanke GT, Kimata-Ariga Y, Standley DM, Kubo A, Taniguchi I, Nakamura H, Hase T (2006). Higher order structure contributes to specific differences in redox potential and electron transfer efficiency of root and leaf ferredoxins. Biochemistry 45, 14389-14396.
DOI URL |
| [18] |
Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An G (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol 44, 463-472.
DOI URL |
| [19] |
Li CM, Hu Y, Huang R, Ma XZ, Wang Y, Liao TT, Zhong P, Xiao FL, Sun CH, Xu ZJ, Deng XJ, Wang PR (2015). Mutation of FdC2 gene encoding a ferredoxin-like protein with C-terminal extension causes yellow-green leaf phenotype in rice. Plant Sci 238, 127-134.
DOI URL |
| [20] | Lichtenthaler HK (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148, 350-382. |
| [21] |
Parks BM, Quail PH (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3, 1177-1186.
DOI URL |
| [22] |
Wang PY, Li CM, Wang Y, Huang R, Sun CH, Xu ZJ, Zhu JQ, Gao XL, Deng XJ, Wang PR (2014). Identification of a geranylgeranyl reductase gene for chlorophyll synthesis in rice. Springerplus 3, 201.
DOI URL |
| [23] |
Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145, 29-40.
DOI URL |
| [24] | Yoo SC, Cho SH, Sugimoto H, Li JJ, Kusumi K, Koh HJ, Iba K, Paek NC (2009). Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150, 388-401. |
| [25] |
Zeng DC, Liu TL, Ma XL, Wang B, Zheng ZY, Zhang YL, Xie XR, Yang BW, Zhao Z, Zhu QL, Liu YG (2020a). Quantitative regulation of Waxy expression by CRISPR/ Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnol J 18, 2385-2387.
DOI URL |
| [26] |
Zeng ZQ, Lin TZ, Zhao JY, Zheng TH, Xu LF, Wang YH, Liu LL, Jiang L, Chen SH, Wan JM (2020b). OsHemA gene, encoding glutamyl-tRNA reductase (GluTR) is essential for chlorophyll biosynthesis in rice (Oryza sativa). J Integr Agricul 19, 612-623.
DOI URL |
| [27] |
Zhang HT, Li JJ, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC (2006). Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62, 325-337.
DOI URL |
| [28] | Zhao J, Qiu ZN, Ruan BP, Kang SJ, Lei H, Zhang S, Dong GJ, Jiang H, Zeng DL, Zhang GH, Gao ZY, Ren DY, Hu XM, Chen G, Guo LB, Qian Q, Zhu L (2015). Functional inactivation of putative photosynthetic electron acceptor ferredoxin C2 (FdC2) induces delayed heading date and decreased photosynthetic rate in rice. PLoS One 10, e0143-361. |
| [29] |
Zhu XY, Guo S, Wang ZW, Du Q, Xing YD, Zhang TQ, Shen WQ, Sang XC, Ling YH, He GH (2016). Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol 16, 134.
DOI URL |
| [1] | Jiahang Che, Weinan Li, Yingzhi Qin, Jinhuan Chen. Research Progress of Leaf Color Variation Mechanism in Woody Plants [J]. Chinese Bulletin of Botany, 2024, 59(2): 319-328. |
| [2] | WANG Ni, LI Zhao-Na, ZHENG Xu-Li, JIANG Si-Cheng, YANG Hai-Yun. Pigment synthesis and photosynthetic characteristics of leaves in Pseudosasa japonica f. akebonosuji [J]. Chin J Plant Ecol, 2024, 48(11): 1536-1546. |
| [3] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [4] | Jiayi Jin, Yiting Luo, Huimin Yang, Tao Lu, Hanfei Ye, Jiyi Xie, Kexin Wang, Qianyu Chen, Yuan Fang, Yuexing Wang, Yuchun Rao. QTL Mapping and Expression Analysis on Candidate Genes Related to Chlorophyll Content in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 394-403. |
| [5] | Qi Zhao, Jibao Jiang, Zenglu Zhang, Qing Jin, Jiali Li, Jiangping Qiu. Species composition and phylogenetic analysis of earthworms on Hainan Island [J]. Biodiv Sci, 2022, 30(12): 22224-. |
| [6] | Dongmei Li, Weihong Yang, Qingduo Li, Xi Han, Xiuping Song, Hong Pan, Yun Feng. High prevalence and genetic variation of Bartonella species inhabiting the bats in southwestern Yunnan [J]. Biodiv Sci, 2021, 29(9): 1245-1255. |
| [7] | Jiangyuan Shang, Yan Chun, Xueyong Li. Map-based Cloning and Natural Variation Analysis of the PAL3 Gene Controlling Panicle Length in Rice [J]. Chinese Bulletin of Botany, 2021, 56(5): 520-532. |
| [8] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| [9] | Jianhua Jiang, Xiaojing Dang, Wenhao Yao, Mengzhu Hu, Yuting Wang, Changmin Hu, Ying Zhang, Dezheng Wang. Genetic Analysis of Four Stigma Traits with Genic Male Sterile Line in Rice (Oryza sativa) [J]. Chinese Bulletin of Botany, 2021, 56(3): 284-295. |
| [10] | Xifeng Chen,Yaping Liu,Bojun Ma. Design and Practice of a New Teaching Project of the Map-based Cloning Experiment in Genetics [J]. Chinese Bulletin of Botany, 2019, 54(6): 797-803. |
| [11] | Zhixiang Chen, Xueying Yao, R. Downie Stephen, Qizhi Wang. Assembling and analysis of Sanicula orthacantha chloroplast genome [J]. Biodiv Sci, 2019, 27(4): 366-372. |
| [12] | Zihong Chen, Xiaona Yang, Ningjing Sun, Ling Xu, Yuan Zheng, Yuming Yang. Species diversity and vertical distribution characteristics of Metarhizium in Gaoligong Mountains, southwestern China [J]. Biodiv Sci, 2018, 26(12): 1308-1317. |
| [13] | Wang Wenle, Feng Dan, Wu Jinxia, Zhang Zhiguo, Lu Tiegang. Gene Mapping of a Dwarf Gene WLD1 in Rice [J]. Chinese Bulletin of Botany, 2017, 52(1): 54-60. |
| [14] | Yongshu Liang, Junjie Zhou, Wenbin Nan, Dongdong Duan, Hanma Zhang. Progress in Rice Root System Research [J]. Chinese Bulletin of Botany, 2016, 51(1): 98-106. |
| [15] | Yuanyong Gong, Shuqiao Guo, Hongmei Shu, Wanchao Ni, Paerhati·Maimaiti, Xinlian Shen, Peng Xu, Xianggui Zhang, Qi Guo. Analysis of Molecular Evolution and Gene Structure of EPSPS Protein in Plant Shikimate Pathway [J]. Chinese Bulletin of Botany, 2015, 50(3): 295-309. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||