

植物学报 ›› 2022, Vol. 57 ›› Issue (1): 80-89.DOI: 10.11983/CBB21120 cstr: 32102.14.CBB21120
王静文1, 王兴军1,2, 马长乐1, 李膨呈1,2,*( )
)
收稿日期:2021-07-23
接受日期:2021-10-12
出版日期:2022-01-01
发布日期:2022-01-17
通讯作者:
李膨呈
作者简介:* E-mail: lpcsaas@outlook.com基金资助:
Jingwen Wang1, Xingjun Wang1,2, Changle Ma1, Pengcheng Li1,2,*( )
)
Received:2021-07-23
Accepted:2021-10-12
Online:2022-01-01
Published:2022-01-17
Contact:
Pengcheng Li
摘要: 核仁是真核细胞中重要的核结构, 核糖体发生最初在核仁中进行, 该过程涉及一系列复杂的反应, 需要许多核仁相关因子参与。核糖体生物发生出现异常通常引起核仁结构紊乱, 并导致细胞周期阻滞、细胞衰老甚至凋亡。核糖体应激响应机制在哺乳动物细胞中研究得较为深入, 但在植物细胞中尚不明晰。尽管如此, 人们逐渐发现某些植物特有的NAC转录因子家族成员在植物细胞中可能参与包括核糖体应激在内的多种胞内应激响应过程。此外, 前期研究发现生长素系统与核糖体生物合成之间存在一种相互协调机制来调控植物发育。该文结合哺乳动物细胞中已知的核糖体应激响应通路, 探讨植物细胞潜在的核糖体应激机制。
王静文, 王兴军, 马长乐, 李膨呈. 植物核糖体应激响应机制研究进展. 植物学报, 2022, 57(1): 80-89.
Jingwen Wang, Xingjun Wang, Changle Ma, Pengcheng Li. A Review on the Mechanism of Ribosome Stress Response in Plants. Chinese Bulletin of Botany, 2022, 57(1): 80-89.
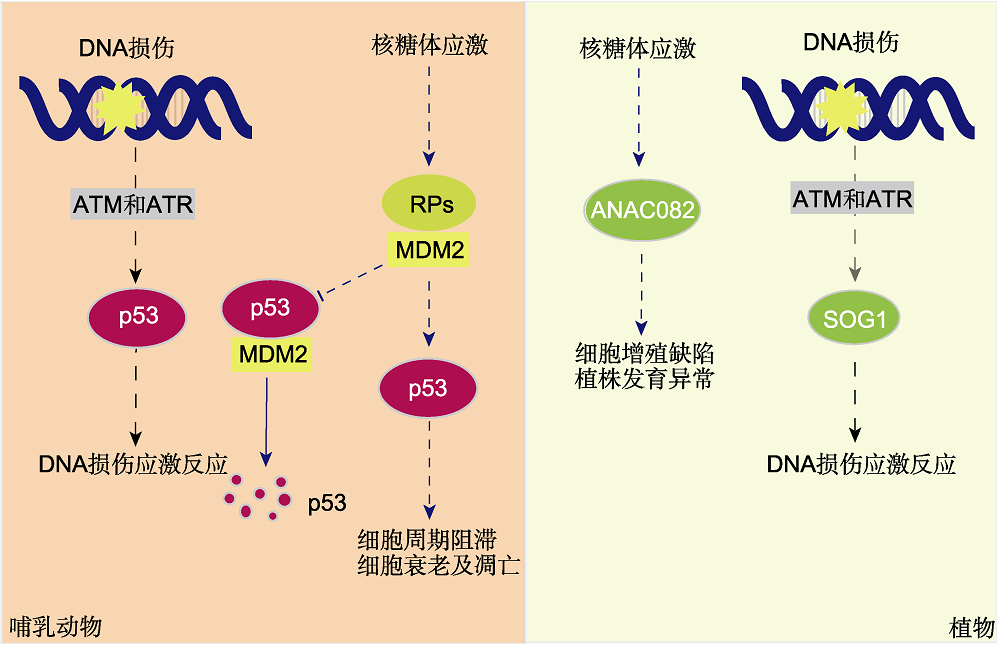
图1 哺乳动物与植物细胞核糖体应激及DNA损伤反应途径比较 核糖体蛋白突变或核糖体生物发生缺陷会导致核糖体应激。在哺乳动物细胞中, 核糖体应激和DNA损伤反应均由p53响应。正常情况下, p53与E3泛素连接酶MDM2相互作用, 通过泛素降解途径调控其蛋白稳态; 当核糖体应激发生时, 一部分RPs从核仁中释放出来, 与MDM2的酸性结构域结合, 降低其对p53的作用。p53作为转录因子介导下游细胞周期阻滞、细胞衰老及凋亡。在植物中, 类p53转录因子SOG1被ATM和ATR激活并磷酸化, 参与DNA损伤反应, 但是目前尚无证据表明SOG1在核糖体应激响应中发挥作用。而另一个NAC家族转录因子ANAC082可能参与一定的核糖体应激反应。实线箭头表示正常状态, 虚线箭头表示应激状态。
Figure 1 Comparison of ribosomal stress and DNA damage response pathways between mammalian and plant cells Ribosomal stress can be resulted from mutations in genes encoding ribosomal proteins or defects in ribosome biogenesis. In mammalian cells, both ribosomal stress and DNA damage responses are mediated by p53. Under normal conditions, p53 interacts with E3 ubiquitin ligase MDM2, which regulates its protein homeostasis through ubiquitin degradation pathway; when ribosomal stress occurs, some RPs are released from the nucleolus and bind to the acidic domain of MDM2, leading to reduction of its effect on p53. p53 acts as a transcription factor to mediate downstream cell cycle arrest, cell aging and apoptosis. In plants, SOG1, a p53-like transcription factor that can be activated and phosphorylated by ATM and ATR, is involved in DNA damage response, but no evidence shows its involvement in ribosomal stress response. However, another NAC transcription factor, ANAC082, is reported to be involved in ribosomal stress response. Solid arrow indicates the normal state, the dotted arrows indicate stress state.
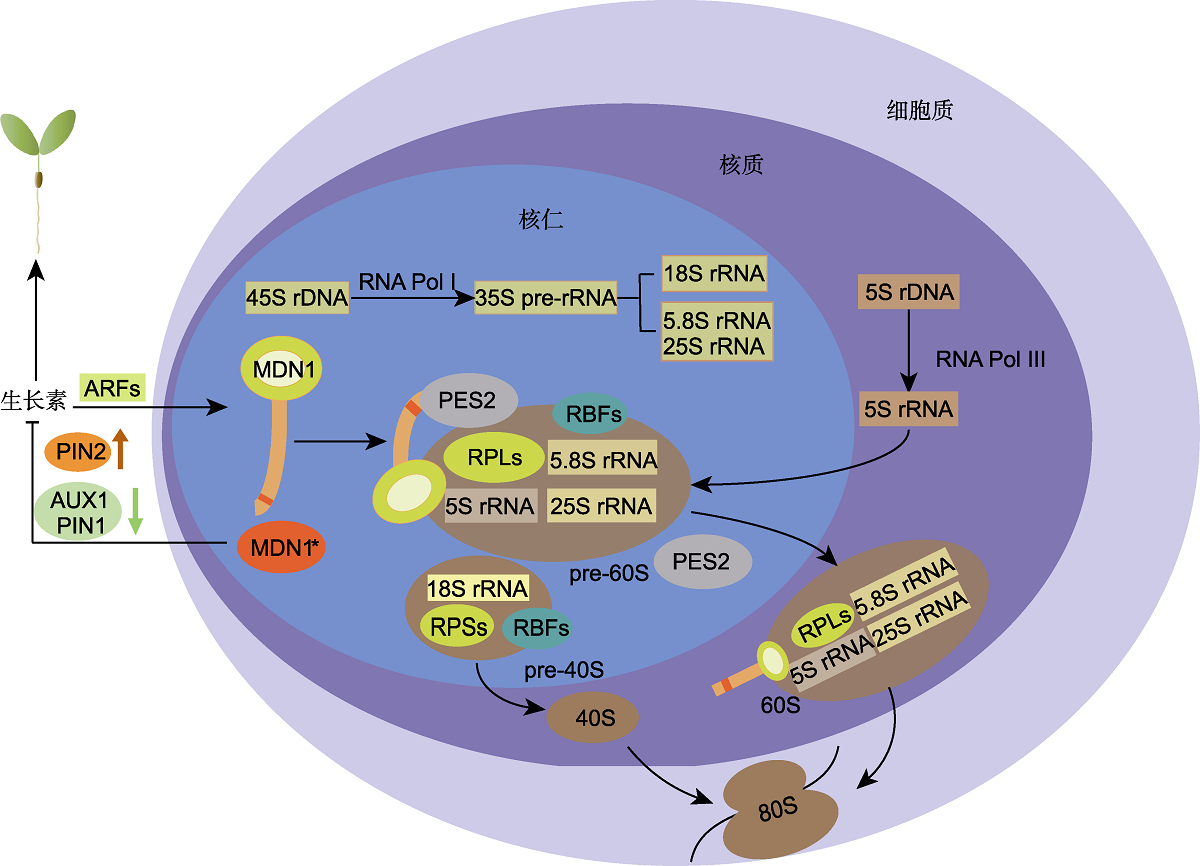
图2 核糖体发生及MDN1相关功能 RNA Pol I: RNA聚合酶I; RPSs/RPLs: 核糖体小/大亚基蛋白; ARFs: 生长素响应因子; RBFs: 核糖体组装因子。在核糖体合成过程中, MDN1与PES2作为60S RBFs参与60S亚基的组装。MDN1的MIDAS结构域能够与PES2的UBL结构域相互作用, 在60S核糖体前体即将进入核质时, MDN1通过“机械力”使PES2从60S前体中解离; 当60S亚基出核进入胞质时, MDN1也从中解离。生长素与核糖体生物发生之间存在相互协调机制, 正常情况下, 生长素能通过ARF激活MDN1的表达; 当MDN1功能发生异常时(用红色MDN1*表示), 会造成PIN2蛋白积累量增加, AUX1和PIN1积累量降低, 进而改变生长素在植株中的稳态和分布。因此, 生长素系统可能通过参与核糖体应激响应来调控植物的生长发育。
Figure 2 Ribosome biogenesis and the related function of MDN1 RNA Pol I: RNA polymerase I; RPSs/RPLs: Ribosomal proteins of the small/large subunit; ARFs: Auxin response factors; RBFs: Ribosomes biogenetic factors. In the process of ribosome biogenesis, both MDN1 and PES2 participate in the assembly of 60S subunits as RBFs. The MIDAS domain of MDN1 can interact with the UBL domain of PES2. When the 60S ribosome precursor is about to enter the nucleoplasm, MDN1 uses ‘mechanical force' to dissociate PES2 from the 60S precursor; at the nuclear export checkpoint, MDN1 is also dissociated from the 60S ribosomal particle. There is a coordination mechanism between auxin and ribosome biogenesis. Under normal conditions, auxin activates the expression of MDN1 through ARFs. When MDN1 is dysfunction (indicated by the red MDN1*), the accumulation of PIN2 protein increases while that of AUX1 and PIN1 decreases, probably leading to changes in both homeostasis and distribution of auxin in plants. Therefore, the auxin system may participate in the ribosomal stress response to regulate plant growth and development.
| [1] |
Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J 64, 960-976.
DOI URL |
| [2] |
Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Söding J, Westhof E, Wilson DN, Beckmann R (2010). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci USA 107, 19748-19753.
DOI URL |
| [3] |
Balazadeh S, Wu AH, Mueller-Roeber B (2010). Salt- triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behav 5, 733-735.
DOI PMID |
| [4] |
Barzilai A, Yamamoto KI (2004). DNA damage responses to oxidative stress. DNA Repair 3, 1109-1115.
PMID |
| [5] |
Baßler J, Hurt E (2019). Eukaryotic ribosome assembly. Annu Rev Biochem 88, 281-306.
DOI URL |
| [6] |
Baßler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010). The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38, 712-721.
DOI URL |
| [7] |
Ben-Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524-1529.
DOI PMID |
| [8] | Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574-585. |
| [9] |
Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010). The nucleolus under stress. Mol Cell 40, 216-227.
DOI URL |
| [10] |
Byrne ME (2009). A role for the ribosome in development. Trends Plant Sci 14, 512-519.
DOI PMID |
| [11] |
Cao J, Geballe AP (1996). Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol 16, 603-608.
DOI PMID |
| [12] |
Cech TT (2000). The ribosome is a ribozyme. Science 289, 878-879.
PMID |
| [13] | Cerezo E, Plisson-Chastang C, Henras AK, Lebaron S, Gleizes PE, O'Donohue MF, Romeo Y, Henry Y (2019). Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip Rev RNA 10, e1516. |
| [14] |
Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS (2011). Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol 31, 4007-4021.
DOI PMID |
| [15] |
Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007). Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029-5037.
PMID |
| [16] |
Clowes FAL (2000). Pattern in root meristem development in angiosperms. New Phytol 146, 83-94.
DOI URL |
| [17] |
Costanzo M, Nishikawa JL, Tang XJ, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004). CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899-913.
PMID |
| [18] | Eichler DC, Craig N (1994). Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49, 197-239. |
| [19] |
Ernst HA, Olsen AN, Skriver K, Larsen S, Lo Leggio L (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5, 297-303.
DOI URL |
| [20] |
Esposito D, Crescenzi E, Sagar V, Loreni F, Russo A, Russo G (2014). Human rpL3 plays a crucial role in cell response to nucleolar stress induced by 5-FU and L-OHP. Oncotarget 5, 11737-11751.
PMID |
| [21] |
Garapati P, Xue GP, Munné-Bosch S, Balazadeh S (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol 168, 1122-1139.
DOI URL |
| [22] |
Gómez-Herreros F, Rodríguez-Galán O, Morillo-Huesca M, Maya D, Arista-Romero M, de la Cruz J, Chávez S, Muñoz-Centeno MC (2013). Balanced production of ribosome components is required for proper G1/S transition in Saccharomyces cerevisiae. J Biol Chem 288, 31689-31700.
DOI PMID |
| [23] |
Grandi P, Rybin V, Baßler J, Petfalski E, Strauß D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E (2002). 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10, 105-115.
PMID |
| [24] |
Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6, 225-242.
DOI URL |
| [25] |
Holmberg Olausson K, Nistér M, Lindström MS (2012). p53-dependent and -independent nucleolar stress re- sponses. Cells 1, 774-798.
DOI PMID |
| [26] |
Horn HF, Vousden KH (2007). Coping with stress: multiple ways to activate p53. Oncogene 26, 1306-1316.
PMID |
| [27] |
Hu WW, Feng ZH, Levine AJ (2012). The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer 3, 199-208.
DOI URL |
| [28] |
James A, Wang YB, Raje H, Rosby R, DiMario P (2014). Nucleolar stress with and without p53. Nucleus 5, 402-426.
DOI URL |
| [29] |
Klinge S, Woolford JL Jr (2019). Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20, 116-131.
DOI URL |
| [30] |
Kornprobst M, Turk M, Kellner N, Cheng JD, Flemming D, Koš-Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R, Hurt E (2016). Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380-393.
DOI PMID |
| [31] |
Koš M, Tollervey D (2010). Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37, 809-820.
DOI URL |
| [32] |
Kressler D, Hurt E, Bergler H, Baßler J (2012). The power of AAA-ATPases on the road of pre-60S ribosome maturation-molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823, 92-100.
DOI PMID |
| [33] |
Kubbutat MHG, Jones SN, Vousden KH (1997). Regulation of p53 stability by Mdm2. Nature 387, 299-303.
DOI URL |
| [34] |
Lafontaine DLJ (2015). Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22, 11-19.
DOI PMID |
| [35] |
Law GL, Raney A, Heusner C, Morris DR (2001). Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276, 38036-38043.
DOI PMID |
| [36] |
Lee MM, Schiefelbein J (1999). WEREWOLF, a MYB- related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473-483.
PMID |
| [37] |
Li K, Zhou XM, Sun XP, Li GH, Hou L, Zhao SZ, Zhao CZ, Ma CL, Li PC, Wang XJ (2021). Coordination between MIDASIN 1-mediated ribosome biogenesis and auxin modulates plant development. J Exp Bot 72, 2501-2513.
DOI URL |
| [38] |
Li PC, Li K, Wang J, Zhao CZ, Zhao SZ, Hou L, Xia H, Ma CL, Wang XJ (2019). The AAA-ATPase MIDASIN 1 functions in ribosome biogenesis and is essential for embryo and root development. Plant Physiol 180, 289-304.
DOI URL |
| [39] |
Lu L, Yi HM, Chen C, Yan SC, Yao H, He GC, Li GF, Jiang YQ, Deng T, Deng XY (2018). Nucleolar stress: is there a reverse version? J Cancer 9, 3723-3727.
DOI PMID |
| [40] |
Maekawa S, Ishida T, Yanagisawa S (2018). Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30, 209-227.
DOI URL |
| [41] |
Mills EW, Green R (2017). Ribosomopathies: there's strength in numbers. Science 358, eaan2755.
DOI URL |
| [42] |
Ohbayashi I, Lin CY, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M (2017). Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29, 2644-2660.
DOI URL |
| [43] | Olson MOJ (2004). Sensing cellular stress: another new function for the nucleolus? Sci STKE 224, pe10. |
| [44] |
Palm D, Streit D, Shanmugam T, Weis BL, Ruprecht M, Simm S, Schleiff E (2019). Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res 47, 1880-1895.
DOI URL |
| [45] |
Pelletier J, Thomas G, Volarević S (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18, 51-63.
DOI PMID |
| [46] |
Petricka JJ, Nelson TM (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol 144, 173-186.
PMID |
| [47] |
Rosado A, Sohn EJ, Drakakaki G, Pan SQ, Swidergal A, Xiong YQ, Kang BH, Bressan RA, Raikhel NV (2010). Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 22, 143-158.
DOI URL |
| [48] |
Sáez-Vásquez J, Delseny M (2019). Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31, 1945-1967.
DOI URL |
| [49] |
Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73, 39-85.
PMID |
| [50] |
Shiloh Y (2001). ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 11, 71-77.
DOI PMID |
| [51] |
Shinohara N, Ohbayashi I, Sugiyama M (2014). Involvement of rRNA biosynthesis in the regulation of CUC1 gene expression and pre-meristematic cell mound formation during shoot regeneration. Front Plant Sci 5, 159.
DOI PMID |
| [52] |
Szakonyi D, Byrne ME (2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65, 269-281.
DOI URL |
| [53] |
Takahashi H, Takahashi A, Naito S, Onouchi H (2012). BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28, 2231-2241.
DOI PMID |
| [54] |
Tomecki R, Sikorski PJ, Zakrzewska-Placzek M (2017). Comparison of preribosomal RNA processing pathways in yeast, plant and human cells-focus on coordinated action of endo- and exoribonucleases. FEBS Lett 591, 1801-1850.
DOI PMID |
| [55] |
Unfried I, Gruendler P (1990). Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18, 4011.
PMID |
| [56] |
Unfried I, Stocker U, Gruendler P (1989). Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Res 17, 7513.
PMID |
| [57] |
Wang WJ, Ryu KH, Bruex A, Barron C, Schiefelbein J (2020). Molecular basis for a cell fate switch in response to impaired ribosome biogenesis in the Arabidopsis root epidermis. Plant Cell 32, 2402-2423.
DOI URL |
| [58] |
Weis BL, Kovacevic J, Missbach S, Schleiff E (2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci 20, 729-740.
DOI URL |
| [59] |
Weis BL, Palm D, Missbach S, Bohnsack MT, Schleiff E (2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21, 415-425.
DOI URL |
| [60] |
Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009). Suppressor of gamma response 1 ( SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106, 12843-12848.
DOI URL |
| [61] |
Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M (2013). ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 14, 817-822.
DOI PMID |
| [62] |
Yusupova G, Yusupov M (2014). High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem 83, 467-486.
DOI PMID |
| [63] |
Zemp I, Kutay U (2007). Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581, 2783-2793.
PMID |
| [1] | 周玉滢, 陈辉, 刘斯穆. 植物非典型Aux/IAA蛋白应答生长素研究进展[J]. 植物学报, 2024, 59(4): 651-658. |
| [2] | 孔祥培, 张蒙悦, 丁兆军. 柳暗花明:胞外生长素信号感受的新突破[J]. 植物学报, 2023, 58(6): 861-865. |
| [3] | 园园, 恩和巴雅尔, 齐艳华. 植物GH3基因家族生物学功能研究进展[J]. 植物学报, 2023, 58(5): 770-782. |
| [4] | 周淑瑶, 李建明, 毛娟. AtGH3.17调控拟南芥生长素和油菜素甾醇的响应[J]. 植物学报, 2023, 58(3): 373-384. |
| [5] | 叶青, 闫晓燕, 陈慧泽, 冯金林, 韩榕. 氮掺杂石墨烯量子点对拟南芥主根生长方向的影响[J]. 植物学报, 2022, 57(5): 623-634. |
| [6] | 王豫颖, 王威浩. 核糖体图谱技术在植物学研究中的应用[J]. 植物学报, 2022, 57(5): 673-683. |
| [7] | 贾利霞, 齐艳华. 生长素代谢、运输及信号转导调控水稻粒型研究进展[J]. 植物学报, 2022, 57(3): 263-275. |
| [8] | 李彬琪, 闫佳慧, 李豪, 辛伟, 田云鹤, 杨贞标, 唐文鑫. 黄瓜卷须缠绕过程中小G蛋白活性变化[J]. 植物学报, 2022, 57(3): 299-307. |
| [9] | 李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41. |
| [10] | 林雨晴, 齐艳华. 生长素输出载体PIN家族研究进展[J]. 植物学报, 2021, 56(2): 151-165. |
| [11] | 黄荣峰, 徐通达. 生长素通过MAPK介导的超长链脂肪酸合成调控侧根发育[J]. 植物学报, 2021, 56(1): 6-9. |
| [12] | 张雨, 赵明洁, 张蔚. 植物次生细胞壁生物合成的转录调控网络[J]. 植物学报, 2020, 55(3): 351-368. |
| [13] | 姚玉婷,马家琦,冯晓莉,潘建伟,王超. 磷酸肌醇激酶FAB1调控拟南芥根毛伸长[J]. 植物学报, 2020, 55(2): 126-136. |
| [14] | 贺祯媚,李东明,齐艳华. 植物ABCB亚家族生物学功能研究进展[J]. 植物学报, 2019, 54(6): 688-698. |
| [15] | 张淑辉,王红,王文茹,吴雪莲,肖元松,彭福田. 蔗糖对桃幼苗生长发育及其SnRK1酶活性的影响[J]. 植物学报, 2019, 54(6): 744-752. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||