

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (3): 285-287.DOI: 10.11983/CBB19060
• COMMENTARIES • Next Articles
Received:2019-03-31
Accepted:2019-04-02
Online:2019-07-01
Published:2019-05-20
Contact:
Ertao Wang
Xiaolin Wang,Ertao Wang. NRT1.1B Connects Root Microbiota and Nitrogen Use in Rice[J]. Chinese Bulletin of Botany, 2019, 54(3): 285-287.
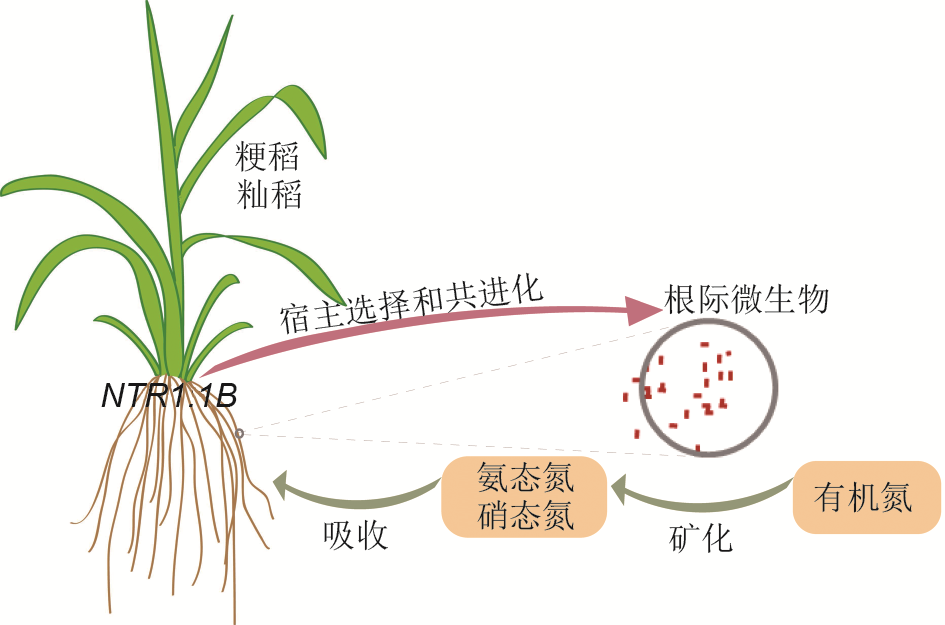
Figure 1 Rice plants coordinate root microbiota to utilize soil nitrogen by NRT1.1B Indica and japonica rice varieties recruit distinct root microb- iota. The biological progresses related to nitrogen metabolism are specifically enriched in indica-enriched bacteria. These bacteria transform organic nitrogen to ammonium, facilitating the nitrogen utilization in rice. NRT1.1B is associated with the recruitments of indica and japonica-specific bacterial taxa.
| [1] |
Beckers B, Op De Beeck M, Weyens N, Van Acker R, Van Montagu M, Boerjan W, Vangronsveld J ( 2016). Lignin engineering in field-grown poplar trees affects the endosphere bacterial microbiome. Proc Natl Acad Sci USA 113, 2312-2317.
DOI URL PMID |
| [2] |
Bender SF, Wagg C, van der Heijden MGA ( 2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31, 440-452.
DOI URL PMID |
| [3] |
Berendsen RL, Pieterse CM, Bakker PA ( 2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17, 478-486.
DOI URL PMID |
| [4] |
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P ( 2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91-95.
DOI URL PMID |
| [5] |
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P ( 2013). Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64, 807-838.
DOI URL PMID |
| [6] |
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM ( 2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fert Soils 48, 489-499.
DOI URL |
| [7] |
Duan L, Liu HB, Li XH, Xiao JH, Wang SP ( 2014). Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant 152, 486-500.
DOI URL PMID |
| [8] |
Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V ( 2015). Structure, variation, and assembly of the root- associated microbiomes of rice. Proc Natl Acad Sci USA 112, E911-E920.
DOI URL PMID |
| [9] |
Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC ( 2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47, 834-838.
DOI URL PMID |
| [10] |
Mbodj D, Effa-Effa B, Kane A, Manneh B, Gantet P, Laplaze L, Diedhiou AG, Grondin A ( 2018). Arbuscular mycorrhizal symbiosis in rice: establishment, environmental control and impact on plant growth and resistance to abiotic stresses. Rhizosphere 8, 12-26.
DOI URL |
| [11] |
Oldroyd GED, Murray JD, Poole PS, Downie JA ( 2011). The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45, 119-144.
DOI URL PMID |
| [12] |
Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT ( 2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA 112, E5013-E5020.
DOI URL PMID |
| [13] |
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET ( 2018). Core microbiomes for sustainable agroecosystems. Nat Plants 4, 247-257.
DOI URL |
| [14] |
Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, Gonzalez-Pena A, Peiffer J, Koren O, Shi Q, Knight R, Glavina del Rio T, Tringe SG, Buckler ES, Dangl JL, Ley RE ( 2018). Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc Natl Acad Sci USA 115, 7368-7373.
DOI URL |
| [15] | Zhang J, Liu Y, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J, Cao S, Wang X, Wang C, Wang H, Qu B, Fan GY, Yuan L, Garrido-Oter R, Chu C, Bai Y ( 2019). NRT1.1B contributes the association of root microbiota and nitrogen use in rice. Nat Biotechnol doi: https://doi.org/10.1038/s41587-019-0104-4. |
| [1] | Xue-Qi GENG YA-KUN TANG WANG LiNa Xu DENG Ze-ling ZHANG Ying ZHOU. Nitrogen addition increases the biomass of Chinese terrestrial plants but reduces their Nitrogen use efficiency [J]. Chin J Plant Ecol, 2024, 48(预发表): 0-0. |
| [2] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. Oryza sativa CYTOKININ OXIDASE 9(OsCKX9)Is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 0-0. |
| [3] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 0-0. |
| [4] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [5] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [6] | Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes [J]. Chinese Bulletin of Botany, 2023, 58(3): 433-439. |
| [7] | Jiayi Jin, Yiting Luo, Huimin Yang, Tao Lu, Hanfei Ye, Jiyi Xie, Kexin Wang, Qianyu Chen, Yuan Fang, Yuexing Wang, Yuchun Rao. QTL Mapping and Expression Analysis on Candidate Genes Related to Chlorophyll Content in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 394-403. |
| [8] | Yuping Yan, Xiaoqi Yu, Deyong Ren, Qian Qian. Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 359-372. |
| [9] | Yuqiang Liu, Jianmin Wan. The Host Controls the Protein Level of Insect Effectors to Balance Immunity and Growth [J]. Chinese Bulletin of Botany, 2023, 58(3): 353-355. |
| [10] | Jingjing Zhao, Haibin Jia, Tien Ming Lee. Market status and the sustainable utilization strategy of wild earthworm (earth dragon) for medicinal use [J]. Biodiv Sci, 2023, 31(3): 22478-. |
| [11] | Qi Wang, Yunzhe Wu, Xueying Liu, Lili Sun, Hong Liao, Xiangdong Fu. The Rice Receptor-like Kinases Function as Key Regulators of Plant Development and Adaptation to the Environment [J]. Chinese Bulletin of Botany, 2023, 58(2): 199-213. |
| [12] | FENG Xu-Fei, LEI Zhang-Ying, ZHANG Yu-Jie, XIANG Dao, YANG Ming-Feng, ZHANG Wang-Feng, ZHANG Ya-Li. Effect of leaf nitrogen allocation on photosynthetic nitrogen use efficiency at flowering and boll stage of Gossypium spp. [J]. Chin J Plant Ecol, 2023, 47(11): 1600-1610. |
| [13] | Liu Xiaolong, Ji Ping, Yang Hongtao, Ding Yongdian, Fu Jialing, Liang Jiangxia, Yu Congcong. Priming Effect of Abscisic Acid on High Temperature Stress During Rice Heading-flowering Stage [J]. Chinese Bulletin of Botany, 2022, 57(5): 596-610. |
| [14] | Wei Heping, Lu Tao, Jia Qiwei, Deng Fei, Zhu Hao, Qi Zehua, Wang Yuxi, Ye Hanfei, Yin Wenjing, Fang Yuan, Mu Dan, Rao Yuchun. QTL Mapping of Candidate Genes for Heading Date in Rice [J]. Chinese Bulletin of Botany, 2022, 57(5): 588-595. |
| [15] | Lixia Jia, Yanhua Qi. Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction [J]. Chinese Bulletin of Botany, 2022, 57(3): 263-275. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
