

植物学报 ›› 2019, Vol. 54 ›› Issue (1): 23-36.DOI: 10.11983/CBB18064 cstr: 32102.14.CBB18064
收稿日期:2018-03-12
接受日期:2018-07-16
出版日期:2019-01-01
发布日期:2019-07-31
通讯作者:
徐凡,张文忠
基金资助:
Xiaoxi Zhen,Haoran Liu,Xin Li,Fan Xu( ),Wenzhong Zhang(
),Wenzhong Zhang( )
)
Received:2018-03-12
Accepted:2018-07-16
Online:2019-01-01
Published:2019-07-31
Contact:
Fan Xu,Wenzhong Zhang
摘要: 氮素是参与植物生长发育的一种重要元素, 对植物的产量和品质具有重要作用。自噬是真核生物中一种保守的细胞组分降解-循环再利用途径, 在植物生长发育和籽粒形成期间的氮素再动员过程中发挥作用。我们鉴定到水稻(Oryza sativa)自噬核心基因OsATG8b, 并获得2个独立的35S-OsATG8b转基因拟南芥(Arabidopsis thaliana)纯合株系。研究表明OsATG8b基因响应低氮胁迫处理, 过表达OsATG8b基因促进转基因拟南芥的生长发育, 使莲座叶增大, 单株产量显著提高(15.16%)。进一步研究表明, 过表达OsATG8b能够显著增强缺氮胁迫下转基因拟南芥叶片中的自噬活性, 从而有效缓解氮胁迫和碳胁迫对转基因拟南芥造成的生长抑制。因此, OsATG8b是提高氮素利用效率和产量的候选基因。
甄晓溪,刘浩然,李鑫,徐凡,张文忠. 异源过表达OsATG8b基因提高转基因拟南芥的 氮/碳胁迫耐受性和产量. 植物学报, 2019, 54(1): 23-36.
Xiaoxi Zhen,Haoran Liu,Xin Li,Fan Xu,Wenzhong Zhang. Heterologous Overexpression of Autophagy-related Gene OsATG8b from Rice Confers Tolerance to Nitrogen/Carbon Starvation and Increases Yield in Arabidopsis. Chinese Bulletin of Botany, 2019, 54(1): 23-36.
| Primer name | Sequence (5′-3′) | Function |
|---|---|---|
| cOsATG8b-F | CCATTCAAGTGGATGGCCAAGAGCTCGTTCAAGC | Gene cloning |
| cOsATG8b-R | GGTGACCTAGAGCAGCCCAAAGGTGTTCTCG | Gene cloning |
| cpOsATG8b-F | AAGCTTAAAATTAAATAAGACGAACAGTCAAACG | Gene cloning |
| cpOsATG8b-R | CCATGGCGCTCCTTCCTGCACACAAT | Gene cloning |
| rtOsATG8b-F | GCTGATCTTACCGTTGGGCA | Real-time RT-PCR |
| rtOsATG8b-R | ATCAGAGCAGCTGTTGGTGG | Real-time RT-PCR |
| rtAtAMT1-F | GCCTCTGCTGACTACTCCAACTT | Real-time RT-PCR |
| rtAtAMT1-R | GACCAGAACCAGTGAGAGACGA | Real-time RT-PCR |
| rtAtNR1-F | AGGATGGGCTAGTAAGCATAAGG | Real-time RT-PCR |
| rtAtNR1-R | GCAAACTGAATCATAGGCGGTG | Real-time RT-PCR |
| rtAtGS2-F | CACCAAACCTTACTCTCTGACA | Real-time RT-PCR |
| rtAtGS2-R | CACTATCTTCACCAGGTGCTTG | Real-time RT-PCR |
| rtAtGDH1-F | GCTTTAGCAGCAACAAACAGAA | Real-time RT-PCR |
| rtAtGDH1-R | TGAGCCAATGCGTTCACTTC | Real-time RT-PCR |
| rtACTIN1-F | ACCATTGGTGCTGAGCGTTT | Real-time RT-PCR |
| rtACTIN1-R | CGCAGCTTCCATTCCTATGAA | Real-time RT-PCR |
| rtTIP41-F | GTATGAAGATGAACTGGCTGACAAT | Real-time RT-PCR |
| rtTIP41-R | ATCAACTCTCAGCCAAAATCGCAAG | Real-time RT-PCR |
表1 引物信息
Table 1 The information of primers
| Primer name | Sequence (5′-3′) | Function |
|---|---|---|
| cOsATG8b-F | CCATTCAAGTGGATGGCCAAGAGCTCGTTCAAGC | Gene cloning |
| cOsATG8b-R | GGTGACCTAGAGCAGCCCAAAGGTGTTCTCG | Gene cloning |
| cpOsATG8b-F | AAGCTTAAAATTAAATAAGACGAACAGTCAAACG | Gene cloning |
| cpOsATG8b-R | CCATGGCGCTCCTTCCTGCACACAAT | Gene cloning |
| rtOsATG8b-F | GCTGATCTTACCGTTGGGCA | Real-time RT-PCR |
| rtOsATG8b-R | ATCAGAGCAGCTGTTGGTGG | Real-time RT-PCR |
| rtAtAMT1-F | GCCTCTGCTGACTACTCCAACTT | Real-time RT-PCR |
| rtAtAMT1-R | GACCAGAACCAGTGAGAGACGA | Real-time RT-PCR |
| rtAtNR1-F | AGGATGGGCTAGTAAGCATAAGG | Real-time RT-PCR |
| rtAtNR1-R | GCAAACTGAATCATAGGCGGTG | Real-time RT-PCR |
| rtAtGS2-F | CACCAAACCTTACTCTCTGACA | Real-time RT-PCR |
| rtAtGS2-R | CACTATCTTCACCAGGTGCTTG | Real-time RT-PCR |
| rtAtGDH1-F | GCTTTAGCAGCAACAAACAGAA | Real-time RT-PCR |
| rtAtGDH1-R | TGAGCCAATGCGTTCACTTC | Real-time RT-PCR |
| rtACTIN1-F | ACCATTGGTGCTGAGCGTTT | Real-time RT-PCR |
| rtACTIN1-R | CGCAGCTTCCATTCCTATGAA | Real-time RT-PCR |
| rtTIP41-F | GTATGAAGATGAACTGGCTGACAAT | Real-time RT-PCR |
| rtTIP41-R | ATCAACTCTCAGCCAAAATCGCAAG | Real-time RT-PCR |
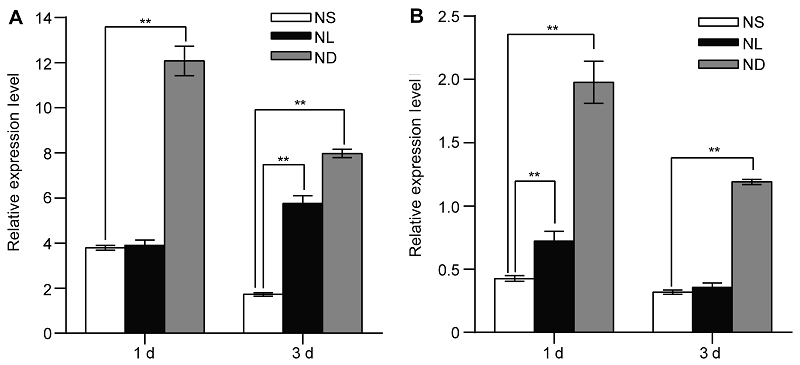
图1 氮胁迫处理诱导水稻幼苗叶片及根中OsATG8b基因的表达 (A) 全氮条件下生长14天的水稻幼苗转移至全氮(NS)、低氮(NL)和缺氮(ND)条件下生长1和3天时叶片中OsATG8b基因的表达量; (B) 全氮、低氮和缺氮条件下生长1和3天的水稻根系中OsATG8b基因的表达量。图中数据为平均值±标准差, n=16, 实验经3次生物学重复。**表示全氮条件与低氮和缺氮条件相比差异显著(P<0.01) (Student’s t-test)。
Figure 1 Identification of OsATG8b as a nitrogen deficiency inducible/responsive gene in leaves and roots of rice seedlings (A) The rice seedlings cultured with N-sufficient (NS) solution for 14 days and transferred to the same NS solution, low N (NL) solution and the N-deficient (ND) solution, the expression of OsATG8b gene in leaves after 1 day and 3 days treatment; (B) The expression of OsATG8b gene in roots after 1 day and 3 days treatment. Values are means±SD, n=16, three biological replicates were performed. ** indicate significant differences in NS solution compared with NL and ND solution (P<0.01) (Student’s t-test).
| WT | L-13 | L-14 | |
|---|---|---|---|
| Bloting time (d) | 36.56±1.58 | 30.78±2.07** | 31.39±1.91** |
| Flowering time (d) | 42.67±1.75 | 35.94±1.98** | 36.61±1.94** |
表2 野生型和35S-OsATG8b转基因拟南芥的抽薹及开花 时间
Table 2 Bolting and flowering times of the wild-type and 35S-OsATG8b transgenic Arabidopsis
| WT | L-13 | L-14 | |
|---|---|---|---|
| Bloting time (d) | 36.56±1.58 | 30.78±2.07** | 31.39±1.91** |
| Flowering time (d) | 42.67±1.75 | 35.94±1.98** | 36.61±1.94** |
| Total number of siliques | Yield per plant (mg) | Thousand grain weight (mg) | |
|---|---|---|---|
| WT | 35.74±3.86 | 85.34±7.89 | 14.87±0.23 |
| L-13 | 46.26±3.13** | 100.13±6.02** | 16.36±0.21** |
| L-14 | 48.22±3.62** | 99.77±5.76** | 17.54±0.41** |
表3 野生型和35S-OsATG8b转基因拟南芥的产量性状
Table 3 Yield related characteristics of the wild-type and 35S-OsATG8b transgenic Arabidopsis
| Total number of siliques | Yield per plant (mg) | Thousand grain weight (mg) | |
|---|---|---|---|
| WT | 35.74±3.86 | 85.34±7.89 | 14.87±0.23 |
| L-13 | 46.26±3.13** | 100.13±6.02** | 16.36±0.21** |
| L-14 | 48.22±3.62** | 99.77±5.76** | 17.54±0.41** |
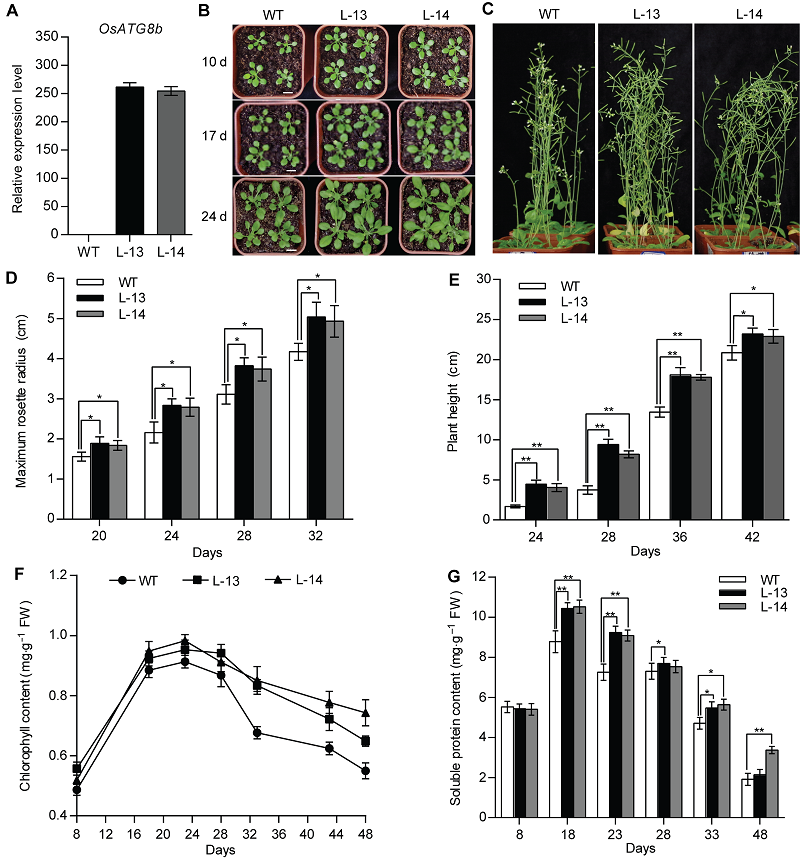
图2 过表达OsATG8b促进转基因拟南芥生长发育 将苗龄8天的野生型(WT)和35S-OsATG8b转基因拟南芥幼苗移入蛭石-营养土(1:3, v/v)中。(A) 苗龄14天的35S-OsATG8b转基因拟南芥和WT幼苗中OsATG8b的表达水平; (B) 由上至下依次为移苗后10、17和24天的拟南芥表型; (C) 移苗后42天的表型; (D) 不同苗龄的35S-OsATG8b转基因拟南芥和WT的最大莲座叶半径; (E) 株高; (F) 总叶绿素含量; (G) 可溶性蛋白含量。Days: 萌发后天数。图中数据为平均值±标准差, n=24, * P<0.05, ** P<0.01 (Student’s t-test), 实验经3次生物学重复。Bars=1 cm
Figure 2 Over-expression of OsATG8b promotes growth and development of transgenic Arabidopsis 8-day-old seedlings were transferred to vermiculite-nutritional soil (1:3, v/v). (A) Expression level of OsATG8b in 14-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT); (B) Panels from top to bottom show phenotypic observations of transgenic lines and WT of Arabidopsis at 10, 17 and 24 days after transfer to soil, respectively; (C) Phenotype of transgenic lines and WT at 42 days after transfer to soil; (D) The maximum rosette radius of 35S-OsATG8b transgenic lines and WT at different seedling age; (E) The plant height; (F) The total chlorophyll content; (G) The soluble protein content. Days: Days after germination. Values are means±SD, n=24, * P<0.05, ** P<0.01 (Student’s t-test), three biological replicates were performed. Bars=1 cm
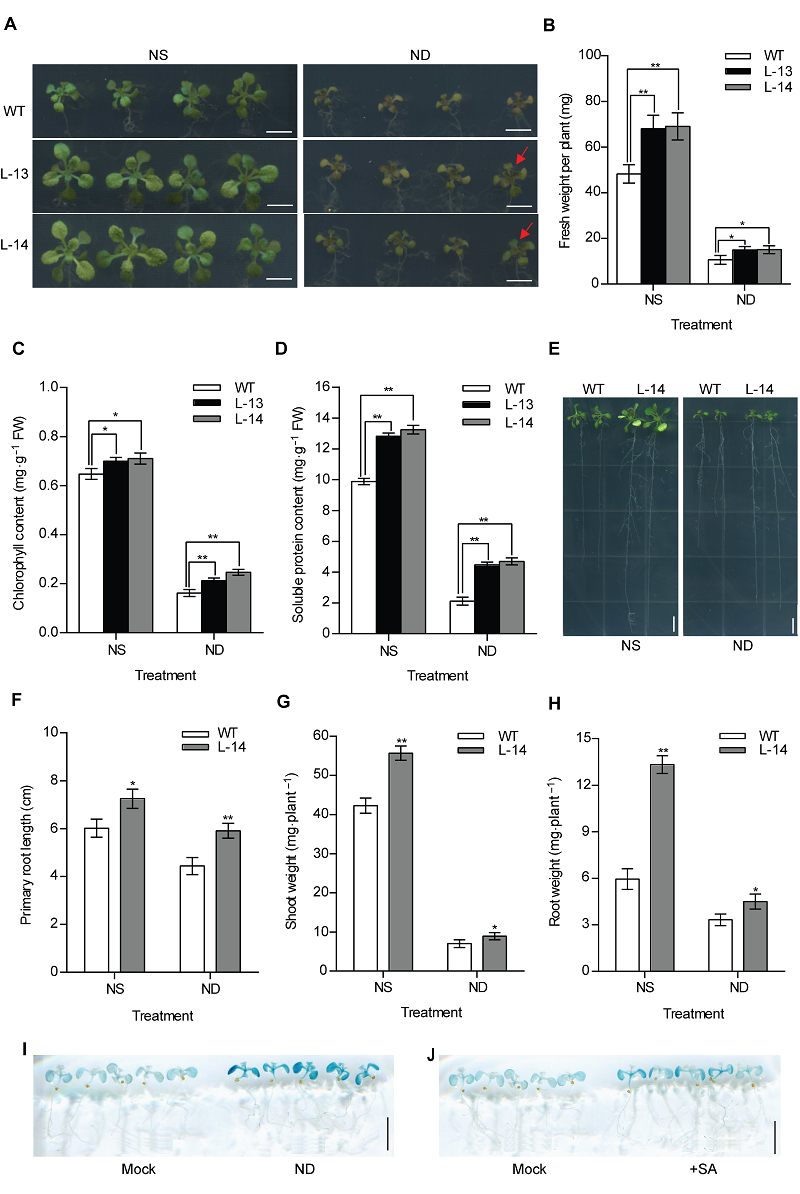
图3 过表达OsATG8b促进转基因拟南芥抵抗缺氮胁迫 (A) 将苗龄7天的35S-OsATG8b转基因和野生型(WT)拟南芥幼苗分别移入全氮(NS)和缺氮(ND)1/2MS培养基中培养9天后的表型; (B)-(D) 分别为全氮(NS)和缺氮(ND)处理9天的35S-OsATG8b转基因和WT拟南芥幼苗的鲜重, 叶绿素和可溶性蛋白含量; (E) 苗龄7天的35S-OsATG8b转基因和WT拟南芥幼苗全氮(NS)和缺氮(ND)垂直培养9天后的表型; (F)-(H) 分别为全氮(NS)和缺氮(ND)处理9天的35S-OsATG8b转基因和WT拟南芥幼苗的主根长、地上部和地下部鲜重; (I), (J) 分别为苗龄10天的ProOsATG8b-GUS转基因拟南芥经缺氮处理和10 μmol·L-1水杨酸(SA)处理24小时后的GUS组织化学染色结果。Mock代表未经处理的拟南芥。数据为平均值±标准差, n=16, *和**分别表示各转基因株系与WT之间差异显著(P<0.05)和极显著(P<0.01) (Student’s t-test), 实验经3次生物学重复。Bars=5 mm
Figure 3 Overexpression of OsATG8b enhances tolerance to N deficiency in transgenic Arabidopsis (A) 7-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT) were transferred to 1/2MS medium for horizontal culture with sufficient (NS) or deficient (ND) N for 9 days. (B)-(D) The fresh weight, chlorophyll content and soluble protein content in rosette leaves of WT and 35S-OsATG8b transgenic Arabidopsis under NS or ND for 9 days, respectively; (E) The phenotype of 7-day-old seedlings of 35S-OsATG8b transgenic lines and WT were transferred to vertical plates with NS or ND for 9 days; (F)-(H) The primary root length, the shoot weight and the root weight of WT and transgenic Arabidopsis lines under NS or ND for 9 days, respectively; (I), (J) 10-day-old seedlings of ProOsATG8b-GUS transgenic Arabidopsis were transferred to ND and 10 μmol·L-1 SA for 24 h, respectively. Mock represented that the seedlings without treated. Values are means±SD, n=16, * and ** indicate significant (P<0.05) and extremely significant (P<0.01) differences between transgenic lines and WT (Student’s t-test), three biological replicates were performed. Bars=5 mm
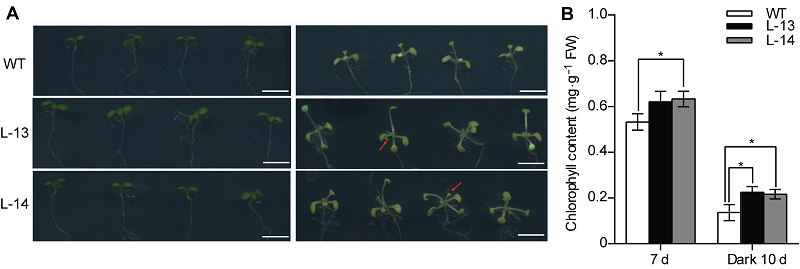
图4 过表达OsATG8b促进转基因拟南芥抵抗黑暗诱导的碳胁迫 (A) 对苗龄7天的35S-OsATG8b转基因拟南芥和野生型(WT)黑暗处理10天后记录表型(左为处理前, 右为处理后); (B) 叶绿素含量测定。数据为平均值±标准差, n=10, *表示各转基因株系与WT之间差异显著(P<0.05) (Student’s t-test), 实验经3次生物学重复。Bars=5 mm
Figure 4 Overexpression of OsATG8b in Arabidopsis enhanced tolerance to carbon starvation induced by dark treatment (A) 7-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT) were transferred to darkness for 10 days (The left is before treatment, and the right is after treatment); (B) The chlorophyll content determination. Values are means±SD, n=10, * indicate significant difference between transgenic lines and WT (P<0.05) (Student’s t-test), three biological replicates were performed. Bars=5 mm
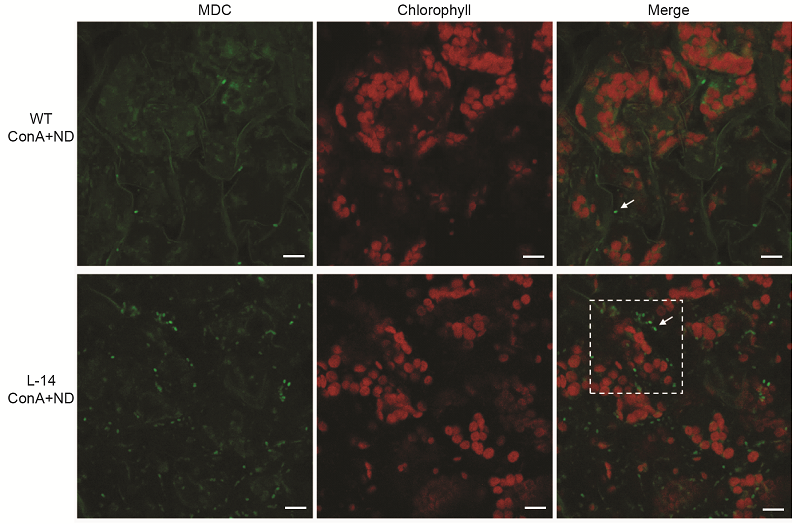
图5 缺氮条件下过表达OsATG8b转基因拟南芥中的自噬活性增加 将苗龄7天的35S-OsATG8b转基因(L-14)和野生型(WT)拟南芥幼苗转移至含有1 μmol·L-1 ConA的缺氮培养液中处理12小时后进行MDC染色, 于激光共聚焦显微镜下观察自噬体荧光。Bars=10 μm
Figure 5 Overexpression of OsATG8b in Arabidopsis enhanced the autophagic activity under N deficient condition 7-day-old seedlings of transgenic line (L-14) and wild type (WT) were transferred to in N-deficient (ND) liquid medium with 1 μmol·L-1 ConA for 12 h, MDC-stained autophagosomes in leaves were observed by confocal microscopy. Bars=10 μm
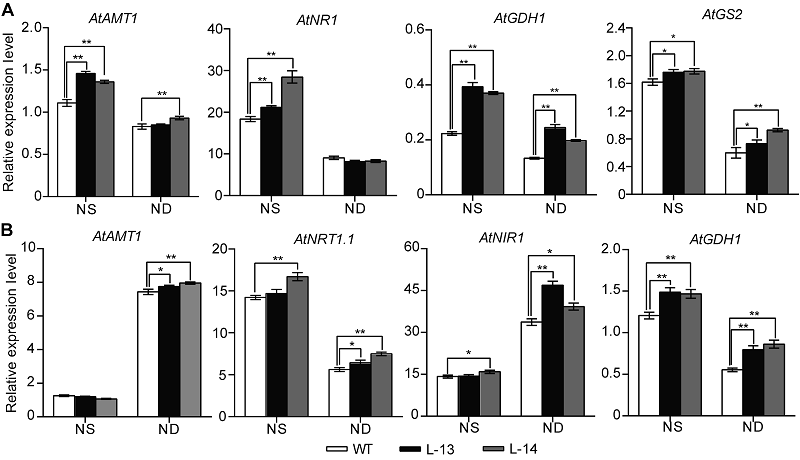
图6 过表达OsATG8b转基因拟南芥中氮代谢关键基因的表达变化 将7天苗龄的35S-OsATG8b转基因和野生型(WT)拟南芥移至全氮(NS)和缺氮(ND) 1/2MS培养基处理14天。(A) 35S-OsATG8b转基因和WT拟南芥中地上部氮代谢相关基因相对表达水平; (B) 地下部氮代谢相关基因相对表达水平。数据为平均值±标准差, n=10, *和**分别表示各转基因株系与WT之间差异显著(P<0.05)和极显著(P<0.01) (Student’s t-test), 实验经3次生物学重复。
Figure 6 Overexpression of OsATG8b in Arabidopsis changes the expression of genes in nitrogen metabolic 7-day-old seedlings of transgenic lines and wild type (WT) were transferred to 1/2MS medium with sufficient (NS) or deficient (ND) nitrogen for 14 days. (A) The expression of genes related to nitrogen metabolic in rosette leaves of 35S-OsATG8b transgenic lines and WT; (B) The expression of genes related to nitrogen metabolic in roots of 35S-OsATG8b transgenic lines and WT. Values are means±SD, n=10, * and ** indicate significant (P<0.05) and extremely significant (P<0.01) differences between transgenic lines and WT (Student’s t-test), respectively, three biological replicates were performed.
| 1 |
黄晓, 李发强 ( 2016). 细胞自噬在植物细胞程序性死亡中的作用. 植物学报 51, 859-862.
DOI URL |
| 2 |
景红娟, 周广舟, 谭晓荣, 平康康, 任雪建 ( 2012). 活性氧对植物自噬调控的研究进展. 植物学报 47, 534-542.
DOI URL |
| 3 | 刘洋, 张静, 王秋玲, 侯岁稳 ( 2018). 植物细胞自噬研究进展. 植物学报 53, 5-16. |
| 4 | 任晨霞, 龚清秋 ( 2014). 细胞自噬在植物碳氮营养中作用的研究进展. 中国细胞生物学学报 36, 407-414. |
| 5 | Arnon DI ( 1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris.Plant Physiol 24, 1-15. |
| 6 | Avila-Ospina L, Moison M, Yoshimoto K, Masclaux- Daubresse C ( 2014). Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65, 3799-3811. |
| 7 |
Biederbick A, Kern HF, Elsässer HP ( 1995). Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles.Eur J Cell Biol 66, 3-14.
DOI URL PMID |
| 8 |
Bradford MM ( 1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248-254.
DOI URL PMID |
| 9 | Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang CJ, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V ( 2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873-894. |
| 10 |
Chardon F, Noël V, Masclaux-Daubresse C ( 2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot 63, 3401-3412.
DOI URL PMID |
| 11 | Clough SJ, Bent AF ( 1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana.Plant J 16, 735-743. |
| 12 |
Contento AL, Xiong Y, Bassham DC ( 2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42, 598-608.
DOI URL PMID |
| 13 | Feng YC, He D, Yao ZY, Klionsky DJ ( 2014). The machi- nery of macroautophagy. Cell Res 24, 24-41. |
| 14 |
Good AG, Shrawat AK, Muench DG ( 2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9, 597-605.
DOI URL PMID |
| 15 | Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux- Daubresse C ( 2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol 199, 683-694. |
| 16 | Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C ( 2012). Autophagy machi- nery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194, 732-740. |
| 17 |
Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T ( 2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process.Plant Physiol 148, 142-155.
DOI URL |
| 18 |
Izumi M, Hidema J, Makino A, Ishida H ( 2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161, 1682-1693.
DOI URL PMID |
| 19 |
Izumi M, Hidema J, Wada S, Kondo E, Kurusu T, Kuchitsu K, Makino A, Ishida H ( 2015). Establishment of monito- ring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol 167, 1307-1320.
DOI URL PMID |
| 20 |
Kichey T, Hirel B, Heumez E, Dubois F, Le Gouis J ( 2007). In winter wheat ( Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers.Field Crops Res 102, 22-32.
DOI URL |
| 21 | Kraiser T, Gras DE, Gutiérrez AG, González B, Gutiérrez RA ( 2011). A holistic view of nitrogen acquisition in plants. J Exp Bot 62, 1455-1466. |
| 22 |
Krapp A ( 2015). Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr Opin Plant Biol 25, 115-122.
DOI URL PMID |
| 23 |
Li FQ, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD ( 2015 a). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389-1408.
DOI URL PMID |
| 24 |
Li WW, Chen M, Wang EH, Hu LQ, Hawkesford MJ, Zhong L, Chen Z, Xu ZS, Li LC, Zhou YB, Guo CH, Ma YZ ( 2016). Genome-wide analysis of autophagy-associated genes in foxtail millet ( Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice.BMC Genomics 17, 797.
DOI URL PMID |
| 25 | Li WW, Chen M, Zhong L, Liu JM, Xu ZS, Li LC, Zhou YB, Guo CH, Ma YZ ( 2015 b). Overexpression of the autophagy-related gene SiATG8a from foxtail millet( Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem Biophys Res Com- mun 468, 800-806. |
| 26 |
Liu D, Gong QQ, Ma YY, Li PL, Li JP, Yang SH, Yuan LL, Yu YQ, Pan DD, Xu F, Wang NN ( 2010). Cpseca, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J Exp Bot 61, 1655-1669.
DOI URL PMID |
| 27 |
Liu YM, Bassham DC ( 2012). Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63, 215-237.
DOI URL PMID |
| 28 |
Makino A, Osmond B ( 1991). Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol 96, 355-362.
DOI URL PMID |
| 29 |
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A ( 2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105, 1141-1157.
DOI URL PMID |
| 30 |
Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M ( 2008). Leaf nitrogen remobilisation for plant development and gr- ain filling. Plant Biol 10, 23-36.
DOI URL PMID |
| 31 | Meyer C, Stitt M ( 2001). Nitrate reduction and signaling. In: Lea PJ, Morot-Gaudry JF, eds. Plant Nitrogen. Berlin, Heidelberg: Springer. pp. 37-59. |
| 32 |
Moriyasu Y, Ohsumi Y ( 1996). Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol 111, 1233-1241.
DOI URL PMID |
| 33 | Ohsumi Y ( 2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2, 211-216. |
| 34 |
Otegui MS, Noh YS, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ ( 2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41, 831-844.
DOI URL PMID |
| 35 |
Patrick JW, Offler CE ( 2001). Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52, 551-564.
DOI URL PMID |
| 36 |
Rentsch D, Schmidt S, Tegeder M ( 2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581, 2281-2289.
DOI URL PMID |
| 37 |
Roberts IN, Caputo C, Criado MV, Funk C ( 2012). Senescence-associated proteases in plants. Physiol Plant 145, 130-139.
DOI URL PMID |
| 38 |
Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G ( 2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot 59, 4029-4043.
DOI URL PMID |
| 39 | Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD ( 2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways.Plant Physiol 138, 2097-2110. |
| 40 | Tsukada M, Ohsumi Y ( 1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae.FEBS Lett 333, 169-174. |
| 41 | Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, Ishida H ( 2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168, 60-73. |
| 42 |
Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A ( 2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149, 885-893.
DOI URL |
| 43 |
Walch-Liu P, Filleur S, Gan YB, Forde BG ( 2005). Signaling mechanisms integrating root and shoot responses to ch- anges in the nitrogen supply. Photosynth Res 83, 239-250.
DOI URL PMID |
| 44 |
Wang P, Sun X, Jia X, Wang N, Gong XQ, Ma FW ( 2016). Characterization of an autophagy-related gene MdATG8i from apple.Front Plant Sci 7, 720.
DOI URL PMID |
| 45 | Wang Y, Yu BJ, Zhao JP, Guo JB, Li Y, Han SJ, Huang L, Du YM, Hong YG, Tang DZ, Liu YL ( 2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383-1399. |
| 46 |
Xia KF, Liu T, Ouyang J, Wang R, Fan T, Zhang MY ( 2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice ( Oryza sativa L.).DNA Res 18, 363-377.
DOI URL PMID |
| 47 | Xia TM, Xiao D, Liu D, Chai WT, Gong QQ, Wang NN ( 2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis.PLoS One 7, e37217. |
| 48 |
Yang XC, Bassham DC ( 2015). New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol 320, 1-40.
DOI URL PMID |
| 49 |
Yao ZY, Delorme-Axford E, Backues SK, Klionsky DJ ( 2015). Atg41/Icy2 regulates autophagosome formation. Autophagy 11, 2288-2299.
DOI URL PMID |
| 50 |
Yoshimoto K ( 2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53, 1355-1365.
DOI URL PMID |
| 51 |
Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y ( 2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967-2983.
DOI URL PMID |
| 52 | Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, OhsumiY, Shirasu K ( 2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914-2927. |
| [1] | 唐远翔, 熊仕臣, 朱洪锋, 张新生, 游成铭, 刘思凝, 谭波, 徐振锋. 长期氮添加对四川盆地西缘常绿阔叶林优势树种凋落叶产量及碳氮磷归还的影响[J]. 植物生态学报, 2025, 49(5): 720-731. |
| [2] | 许庭旸, 刘雨辰, 王万鹏, 苏航, 苏昆龙, 吴振映, 吕明, 李福利, 王小山, 付春祥. 喷施不同植物生长调节剂对盐碱地小麦生长发育的影响[J]. 植物学报, 2025, 60(3): 354-362. |
| [3] | 严语萍, 俞晓琦, 任德勇, 钱前. 水稻穗粒数遗传机制与育种利用[J]. 植物学报, 2023, 58(3): 359-372. |
| [4] | 刘建新, 刘瑞瑞, 刘秀丽, 贾海燕, 卜婷, 李娜. 外源硫化氢对盐碱胁迫下裸燕麦光合碳代谢的调控[J]. 植物生态学报, 2023, 47(3): 374-388. |
| [5] | 刘裕强, 万建民. 寄主监控昆虫唾液蛋白平衡植物抗性与生长发育[J]. 植物学报, 2023, 58(3): 353-355. |
| [6] | 叶卫军, 张阴, 王沛然, 张玲玲, 田东丰, 吴泽江, 周斌. 绿豆5个产量相关性状的QTL分析[J]. 植物学报, 2023, 58(1): 150-158. |
| [7] | 刘晓龙, 季平, 杨洪涛, 丁永电, 付佳玲, 梁江霞, 余聪聪. 脱落酸对水稻抽穗开花期高温胁迫的诱抗效应[J]. 植物学报, 2022, 57(5): 596-610. |
| [8] | 王雷, 种康. 鱼和熊掌的选择: 反向重复序列变异介导的玉米环境适应与产量平衡[J]. 植物学报, 2022, 57(5): 555-558. |
| [9] | 熊淑萍, 曹文博, 曹锐, 张志勇, 付新露, 徐赛俊, 潘虎强, 王小纯, 马新明. 水平结构配置对冬小麦冠层垂直结构、微环境及产量的影响[J]. 植物生态学报, 2022, 46(2): 188-196. |
| [10] | 孙浩哲, 王襄平, 张树斌, 吴鹏, 杨蕾. 阔叶红松林不同演替阶段凋落物产量及其稳定性的影响因素[J]. 植物生态学报, 2021, 45(6): 594-605. |
| [11] | 周俭民. 免疫信号轴揭示水稻与病原菌斗争的秘密[J]. 植物学报, 2021, 56(5): 513-515. |
| [12] | 李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超. 植物细胞自噬基因的功能与转录调控机制[J]. 植物学报, 2021, 56(2): 201-217. |
| [13] | 王泽义, 张恒嘉, 王玉才, 陈谢田, 巴玉春. 亏缺灌溉对板蓝根叶片光合生理特性及产量的影响[J]. 植物学报, 2020, 55(6): 705-714. |
| [14] | 刘雪飞, 吴林, 王涵, 洪柳, 熊莉军. 鄂西南亚高山湿地泥炭藓的生长与分解[J]. 植物生态学报, 2020, 44(3): 228-235. |
| [15] | 张硕, 吴昌银. 长链非编码RNA基因Ef-cd调控水稻早熟与稳产[J]. 植物学报, 2019, 54(5): 550-553. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||