

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (4): 651-658.DOI: 10.11983/CBB23106 cstr: 32102.14.CBB23106
• SPECIAL TOPICS • Previous Articles Next Articles
Yuying Zhou1, Hui Chen2,*( ), Simu Liu1,*(
), Simu Liu1,*( )
)
Received:2023-08-03
Accepted:2023-11-14
Online:2024-07-10
Published:2024-07-10
Contact:
*E-mail: chenhui2@zstp.edu.cn; liusm@szu.edu.cn
Yuying Zhou, Hui Chen, Simu Liu. Research Progress on Auxin Responsive Non-canonical Aux/IAA Proteins in Plants[J]. Chinese Bulletin of Botany, 2024, 59(4): 651-658.
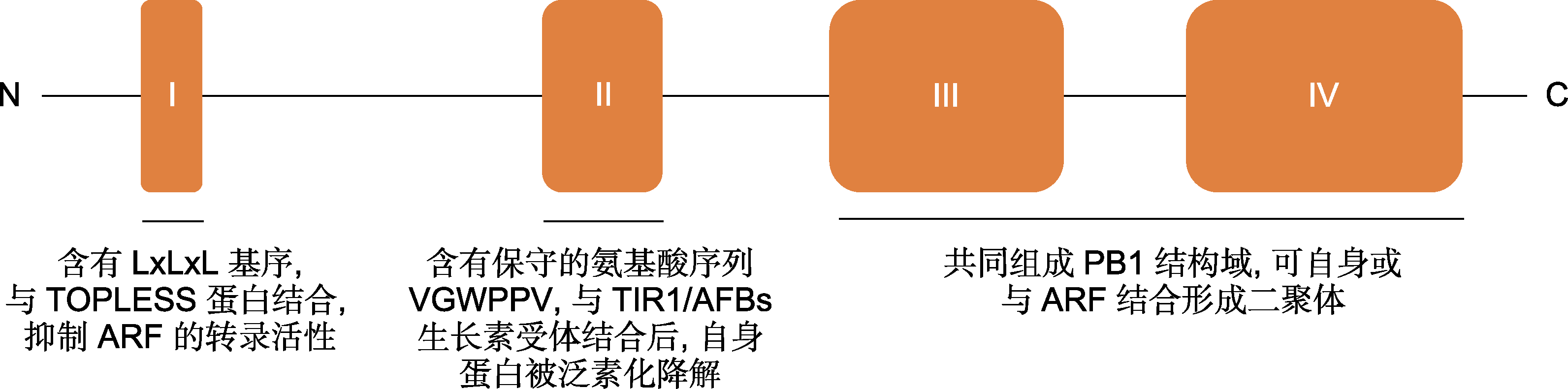
Figure 1 Functional domain diagram of canonical Aux/IAA protein (Tiwari et al., 2004; Szemenyei et al., 2008; Calderón Villalobos et al., 2012; Guilfoyle, 2015)
| 非典型Aux/ IAA蛋白 | 结构域I | 结构域II |
|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | ||
| AtIAA20 | SSSSSSIES | ···SVAAPAVEDAEYV |
| AtIAA30 | SSSSSSIES | ···EYDG··VGAAEEM |
| AtIAA31 | ········· | REARQDWPPIKSRLRD |
| AtIAA32 | NTPADFFKG | ELIDWSQPSYNSITQL |
| AtIAA33 | ········· | SSGAAG·RSFQGFGLN |
| AtIAA34 | PHPLHLVAS | GVIDLG·LSLRTIQHE |
| 水稻 (Oryza sativa) | ||
| OsIAA4 | ········· | ················ |
| OsIAA8 | TDLRLGLTL | VVHIDGNNPSTPRSSL |
| OsIAA26 | TELTLGPPG | RGRKNGHPPPSSSM·· |
| OsIAA27 | ········· | DETTAPPPRSAAAATE |
| OsIAA28 | ········· | DRNASAGPEVKPAGLS |
| OsIAA29 | ········· | DRNASAEPVVKP·GLS |
Table 1 Non-canonical Aux/IAA proteins lack the LxLxL motif of domain I and/or the VGWPPV core amino acid resi- dues of domain II
| 非典型Aux/ IAA蛋白 | 结构域I | 结构域II |
|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | ||
| AtIAA20 | SSSSSSIES | ···SVAAPAVEDAEYV |
| AtIAA30 | SSSSSSIES | ···EYDG··VGAAEEM |
| AtIAA31 | ········· | REARQDWPPIKSRLRD |
| AtIAA32 | NTPADFFKG | ELIDWSQPSYNSITQL |
| AtIAA33 | ········· | SSGAAG·RSFQGFGLN |
| AtIAA34 | PHPLHLVAS | GVIDLG·LSLRTIQHE |
| 水稻 (Oryza sativa) | ||
| OsIAA4 | ········· | ················ |
| OsIAA8 | TDLRLGLTL | VVHIDGNNPSTPRSSL |
| OsIAA26 | TELTLGPPG | RGRKNGHPPPSSSM·· |
| OsIAA27 | ········· | DETTAPPPRSAAAATE |
| OsIAA28 | ········· | DRNASAGPEVKPAGLS |
| OsIAA29 | ········· | DRNASAEPVVKP·GLS |
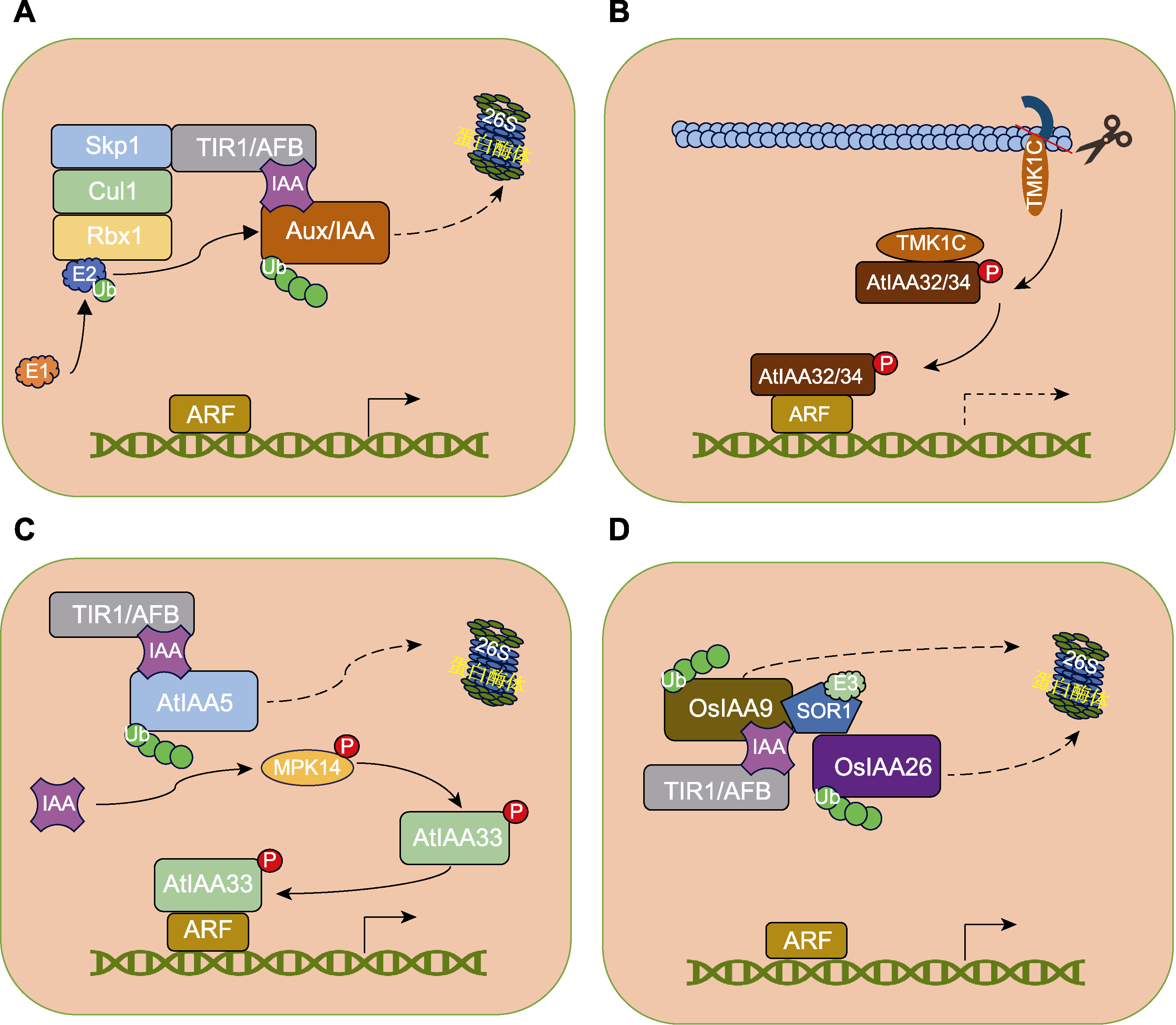
Figure 2 Aux/IAA-mediated auxin signaling pathway (Chen et al., 2018; Mutte et al., 2018; Cao et al., 2019; Lv et al., 2020; Yu et al., 2022) (A) Canonical Aux/IAA-mediated auxin signaling pathway; (B) TMK1-AtIAA32/34-mediated auxin signaling pathway in Arabidopsis; (C) MPK14-AtIAA33-mediated auxin signaling pathway in Arabidopsis; (D) SOR1-OsIAA26-mediated auxin signaling in rice.
| [1] |
Basunia MA, Nonhebel HM, Backhouse D, McMillan M (2021). Localised expression of OsIAA29 suggests a key role for auxin in regulating development of the dorsal aleurone of early rice grains. Planta 254, 40.
DOI PMID |
| [2] | Béziat C, Kleine-Vehn J (2018). The road to auxin-dependent growth repression and promotion in apical hooks. Curr Biol 28, R519-R525. |
| [3] |
Calderón Villalobos LIA, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao HB, Zheng N, Napier R, Kepinski S, Estelle M (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8, 477-485.
DOI PMID |
| [4] | Cao M, Chen R, Li P, Yu YQ, Zheng R, Ge DF, Zheng W, Wang XH, Gu YT, Gelová Z, Friml J, Zhang H, Liu RY, He J, Xu TD (2019). TMK1-mediated auxin signaling regulates differential growth of the apical hook. Nature 568, 240-243. |
| [5] |
Chen H, Ma B, Zhou Y, He SJ, Tang SY, Lu X, Xie Q, Chen SY, Zhang JS (2018). E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc Natl Acad Sci USA 115, 4513- 4518.
DOI PMID |
| [6] | Dharmasiri N, Dharmasiri S, Estelle M (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441-445. |
| [7] | Ding ZJ, Friml J (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107, 12046-12051. |
| [8] |
Dreher KA, Brown J, Saw RE, Callis J (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699-714.
DOI PMID |
| [9] |
Figueiredo MRA, Strader LC (2022). Intrinsic and extrinsic regulators of Aux/IAA protein degradation dynamics. Trends Biochem Sci 47, 865-874.
DOI PMID |
| [10] | Friml J, Gallei M, Gelová Z, Johnson A, Mazur E, Monzer A, Rodriguez L, Roosjen M, Verstraeten I, Živanović BD, Zou MX, Fiedler L, Giannini C, Grones P, Hrtyan M, Kaufmann WA, Kuhn A, Narasimhan M, Randuch M, Rýdza N, Takahashi K, Tan ST, Teplova A, Kinoshita T, Weijers D, Rakusová H (2022). ABP1-TMK auxin perception for global phosphorylation and auxin canalization. Nature 609, 575-581. |
| [11] | Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271-276. |
| [12] | Guilfoyle TJ (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33-43. |
| [13] | Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S, Suh JY (2014). Structural basis for the auxin-induced transcrip-tional regulation by Aux/IAA17. Proc Natl Acad Sci USA 111, 18613-18618. |
| [14] | Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sati-va). Funct Integr Genomics 6, 47-59. |
| [15] | Jing HW, Yang XL, Emenecker RJ, Feng J, Zhang J, Chaisupa P, Wright RC, Holehouse AS, Strader LC, Zuo JR (2023). Nitric oxide-mediated S-nitrosylation of IAA17 protein in intrinsically disordered region represses auxin signaling. J Genet Genomics 50, 473-485. |
| [16] | Jing HW, Yang XL, Zhang J, Liu XH, Zheng HK, Dong GJ, Nian JQ, Feng J, Xia B, Qian Q, Li JY, Zuo JR (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signaling. Nat Com-mun 6, 7395. |
| [17] | Kepinski S, Leyser O (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446-451. |
| [18] | Li LX, Gallei M, Friml J (2022). Bending to auxin: fast acid growth for tropisms. Trends Plant Sci 27, 440-449. |
| [19] | Li YY, Qi YH (2022). Advances in biological functions of Aux/IAA gene family in plants. Chin Bull Bot 57, 30-41. (in Chinese) |
|
李艳艳, 齐艳华 (2022). 植物Aux/IAA基因家族生物学功能研究进展. 植物学报 57, 30-41.
DOI |
|
| [20] | Lin WW, Zhou X, Tang W, Takahashi K, Pan X, Dai JW, Ren H, Zhu XY, Pan SQ, Zheng HY, Gray WM, Xu TD, Kinoshita T, Yang ZB (2021). TMK-based cell-surface auxin signaling activates cell-wall acidification. Nature 599, 278-282. |
| [21] | Lv BS, Yu QQ, Liu JJ, Wen XJ, Yan ZW, Hu KQ, Li HB, Kong XP, Li CL, Tian HY, De Smet I, Zhang XS, Ding ZJ (2020). Non-canonical AUX/IAA protein IAA33 compe-tes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J 39, e101515. |
| [22] |
Mockaitis K, Estelle M (2008). Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24, 55-80.
DOI PMID |
| [23] | Müller CJ, Valdés AE, Wang GD, Ramachandran P, Beste L, Uddenberg D, Carlsbecker A (2016). PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in Arabidopsis. Plant Physiol 170, 956-970. |
| [24] | Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D (2018). Origin and evolution of the nuclear auxin response system. eLife 7, e33399. |
| [25] |
Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang SC, Hagen G, Li HB, Guilfoyle TJ, Parcy F, Vernoux T, Dumas R (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5, 3617.
DOI PMID |
| [26] | Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM (2011). Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol 156, 537-549. |
| [27] | Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T (2014). Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5, 744. |
| [28] |
Reed JW (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6, 420-425.
DOI PMID |
| [29] |
Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371-1373.
DOI PMID |
| [30] | Salehin M, Bagchi R, Estelle M (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27, 9-19. |
| [31] | Sato A, Yamamoto KT (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 133, 397- 405. |
| [32] |
Shimizu-Mitao Y, Kakimoto T (2014). Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55, 1450-1459.
DOI PMID |
| [33] | Song YL, Xu ZF (2013). Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces res-ponsiveness to auxin. Int J Mol Sci 14, 13645-13656. |
| [34] |
Szemenyei H, Hannon M, Long JA (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384-1386.
DOI PMID |
| [35] | Tan X, Calderon-Villalobos LIA, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645. |
| [36] | Teale WD, Paponov IA, Palme K (2006). Auxin in action: signaling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7, 847-859. |
| [37] | Teng ZN, Yu HH, Wang GQ, Meng S, Liu BH, Yi YK, Chen YK, Zheng Q, Liu L, Yang JC, Duan MJ, Zhang JH, Ye NH (2022). Synergistic interaction between ABA and IAA due to moderate soil drying promotes grain filling of infe-rior spikelets in rice. Plant J 109, 1457-1472. |
| [38] |
Tiwari SB, Hagen G, Guilfoyle TJ (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533-543.
DOI PMID |
| [39] |
Vain T, Raggi S, Ferro N, Barange DK, Kieffer M, Ma Q, Doyle SM, Thelander M, Pařízková B, Novák O, Ismail A, Enquist PA, Rigal A, Łangowska M, Ramans Harbo-rough S, Zhang Y, Ljung K, Callis J, Almqvist F, Kepinski S, Estelle M, Pauwels L, Robert S (2019). Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc Natl Acad Sci USA 116, 6463-6472.
DOI PMID |
| [40] |
Wang RH, Estelle M (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol 21, 51-58.
DOI PMID |
| [41] |
Yu ZP, Zhang F, Friml J, Ding ZJ (2022). Auxin signaling: research advances over the past 30 years. J Integr Plant Biol 64, 371-392.
DOI |
| [42] | Zhong SW, Shi H, Xue C, Wei N, Guo HW, Deng XW (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA 111, 3913-3920. |
| [1] |
Liang Ma, Yongqing Yang, Yan Guo.
“Next-generation Green Revolution” Genes: Toward New “Climate-Smart” Crop Breeding [J]. Chinese Bulletin of Botany, 2025, 60(4): 489-498. |
| [2] | Jiayi Feng, Juyu Lian, Yujun Feng, Dongxu Zhang, Honglin Cao, Wanhui Ye. Effects of vertical stratification on community structure and functions in a subtropical, evergreen broad-leaved forest in the Dinghushan National Nature Reserve [J]. Biodiv Sci, 2024, 32(12): 24306-. |
| [3] | Xiangpei Kong, Mengyue Zhang, Zhaojun Ding. There Is a Way Out-new Breakthroughs in Extracellular Auxin Sensing [J]. Chinese Bulletin of Botany, 2023, 58(6): 861-865. |
| [4] | Yuan Yuan, Enhebayaer, Qi Yanhua. Research Advances in Biological Functions of GH3 Gene Family in Plants [J]. Chinese Bulletin of Botany, 2023, 58(5): 770-782. |
| [5] | Shuyao Zhou, Jianming Li, Juan Mao. AtGH3.17-mediated Regulation of Auxin and Brassinosteroid Response in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2023, 58(3): 373-384. |
| [6] | Jiman Li, Nan Jin, Maogang Xu, Jusong Huo, Xiaoyun Chen, Feng Hu, Manqiang Liu. Effects of earthworm on tomato resistance under different drought levels [J]. Biodiv Sci, 2022, 30(7): 21488-. |
| [7] | Ye Qing, Yan Xiaoyan, Chen Huize, Feng Jinlin, Han Rong. Effect of Nitrogen-doped Graphene Quantum Dots on Growth Direction of Primary Root in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2022, 57(5): 623-634. |
| [8] | Lixia Jia, Yanhua Qi. Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction [J]. Chinese Bulletin of Botany, 2022, 57(3): 263-275. |
| [9] | Binqi Li, Jiahui Yan, Hao Li, Wei Xin, Yunhe Tian, Zhenbiao Yang, Wenxin Tang. Changes of Small GTPases Activity During Cucumber Tendril Winding [J]. Chinese Bulletin of Botany, 2022, 57(3): 299-307. |
| [10] | Yanyan Li, Yanhua Qi. Advances in Biological Functions of Aux/IAA Gene Family in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 30-41. |
| [11] | Jingwen Wang, Xingjun Wang, Changle Ma, Pengcheng Li. A Review on the Mechanism of Ribosome Stress Response in Plants [J]. Chinese Bulletin of Botany, 2022, 57(1): 80-89. |
| [12] | Xiaomin Cui, Dongchao Ji, Tong Chen, Shiping Tian. Advances in the Studies on Molecular Mechanism of Receptor-like Protein Kinase FER Regulating Host Plant-pathogen Interaction [J]. Chinese Bulletin of Botany, 2021, 56(3): 339-346. |
| [13] | Yuqing Lin, Yanhua Qi. Advances in Auxin Efflux Carrier PIN Proteins [J]. Chinese Bulletin of Botany, 2021, 56(2): 151-165. |
| [14] | Rongfeng Huang, Tongda Xu. Auxin Regulates the Lateral Root Development Through MAPK-mediated VLCFAs Biosynthesis [J]. Chinese Bulletin of Botany, 2021, 56(1): 6-9. |
| [15] | Chenghuizi Yang,Xianyu Tang,Wei Li,Shitou Xia. NLR and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2020, 55(4): 497-504. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||