

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (1): 44-49.DOI: 10.11983/CBB20203 cstr: 32102.14.CBB20203
• INVITED PROTOCOLS • Previous Articles Next Articles
Xianrong Xie, Dongchang Zeng, Jiantao Tan, Qinlong Zhu, Yaoguang Liu*( )
)
Received:2020-12-11
Accepted:2021-02-25
Online:2021-01-01
Published:2021-02-25
Contact:
Yaoguang Liu
Xianrong Xie, Dongchang Zeng, Jiantao Tan, Qinlong Zhu, Yaoguang Liu. CRISPR-based DNA Fragment Deletion in Plants[J]. Chinese Bulletin of Botany, 2021, 56(1): 44-49.
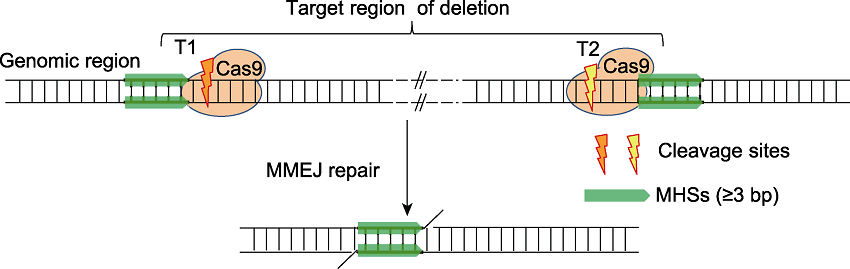
Figure 1 The principle for microhomology-mediated end joining (MMEJ)-based DNA fragment deletion(modified from Xie et al., 2020) MHS: Microhomologous sequence
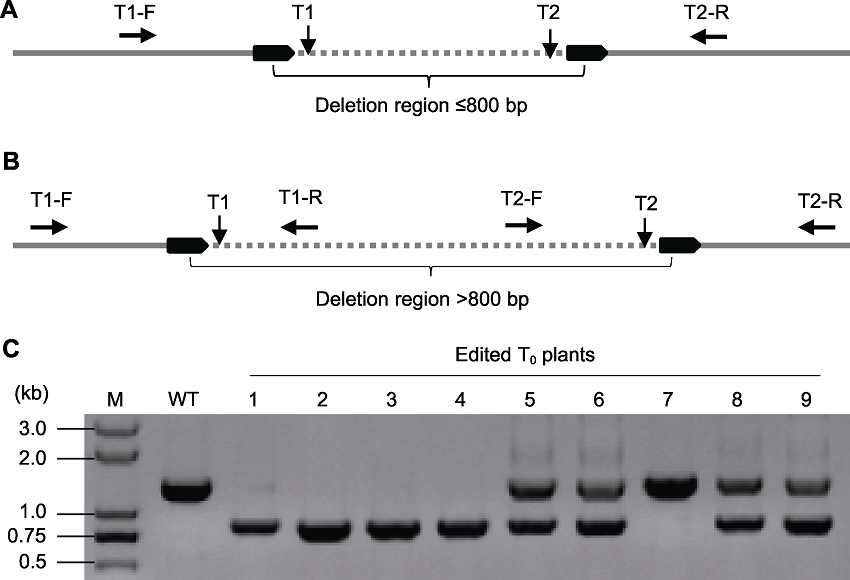
Figure 3 The strategy for identification of the mutant types in transgenic plants (A) Amplification of the target region using the forward primer (T1-F) upstream the T1 and reverse primer (T2-R) downstream T2 if the length of desirable deleted fragment is less than 800 bp; (B) Four primers (with the length of the following amplification products is about 70-100 bp different from each other) with three combinations (T1-F/T1-R, T2-F/T2-R, and T1-F/T2-R) or pooled in an amplification are suggested if the length of desirable deleted fragment is more than 800 bp (there is no deletion of the target region if the resulting bands are produced with primer groups (T1-F/T1-R and T2-F/T2-R) but not T1-F/T2-R; both alleles had fragment deletion if the resulting bands are produced with primer group T1-F/T2-R but not T1-F/T1-R and T2-F/T2-R; one allele had fragment deletion and the other allele had no fragment deletion if all three fragments were produced; (C) The example of gel electrophoresis according to the detection method of (A). WT: Wild type; 1-9 indicate edited T0 plants in which 1-4 with biallelic fragment deletion, 5, 6, 8 and 9 with heterozygous deletion and 7 with no deletion.
| [1] | 苏钺凯, 邱镜仁, 张晗, 宋振巧, 王建华 (2019). CRISPR/ Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报 54, 385-395. |
| [2] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光 (2018). 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法. 中国科学: 生命科学 48, 783-794. |
| [3] |
Burgess DG, Xu J, Freeling M (2015). Advances in understanding cis regulation of the plant gene with an emphasis on comparative genomics. Curr Opin Plant Biol 27, 141-147.
DOI URL |
| [4] |
Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH (2014). Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRIS- PR)/Cas9 nuclease system in mammalian cells. J Biol Chem 289, 21312-21324.
DOI URL PMID |
| [5] | Liu WZ, Xie XR, Ma XL, Li J, Chen JH, Liu YG (2015). DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8, 1431-1433. |
| [6] |
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274-1284.
URL PMID |
| [7] |
Ma XL, Zhu QL, Chen YL, Liu YG (2016). CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant 9, 961-974.
URL PMID |
| [8] |
Manova V, Gruszka D (2015). DNA damage and repair in plants-from models to crops. Front Plant Sci 6, 885.
URL PMID |
| [9] | Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, Codner GF, Hublitz P, McHugh PJ, Teboul L, de Bruijn MFTR (2019). Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47, 7402-7417. |
| [10] |
Tan JT, Zhao YC, Wang B, Hao Y, Wang YX, Li YY, Luo WN, Zong WB, Li GS, Chen SF, Ma K, Xie XR, Chen LT, Liu YG, Zhu QL (2020). Efficient CRISPR/Cas9-based plant genomic fragment deletions by microhomology- mediated end joining. Plant Biotechnol J 18, 2161-2163.
DOI URL PMID |
| [11] |
Tuladhar R, Yeu Y, Piazza JT, Tan ZP, Clemenceau JR, Wu XF, Barrett Q, Herbert J, Mathews DH, Kim J, Hwang TH, Lum L (2019). CRISPR-Cas9-based mu- tagenesis frequently provokes on-target mRNA misregulation. Nat Commun 10, 4056.
URL PMID |
| [12] |
Xie XR, Liu WZ, Dong G, Zhu QH, Liu YG (2020). MMEJ-KO: a web tool for designing paired CRISPR guide RNAs for microhomology-mediated end joining fragment deletion. Sci China Life Sci doi: 10.1007/s11427-020-1797-3.
URL PMID |
| [13] |
Xie XR, Ma XL, Zhu QL, Zeng DC, Li GS, Liu YG (2017). CRISPR-GE: a convenient software toolkit for CRISPR- based genome editing. Mol Plant 10, 1246-1249.
DOI URL PMID |
| [14] | Zhang Y, Massel K, Godwin ID, Gao CX (2018). Applications and potential of genome editing in crop improvement. Genome Biol 19, 210. |
| [1] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [2] | Danling Hu, Yongwei Sun. Advances in Virus-mediated Genome Editing Technology in Plants [J]. Chinese Bulletin of Botany, 2024, 59(3): 452-462. |
| [3] | Chunyan Miao, Mingming Li, Xin Zuo, Ning Ding, Jiafang Du, Juan Li, Zhongyi Zhang, Fengqing Wang. Establishment of CRISPR/Cas9 Gene Editing System in Rehmannia henryi [J]. Chinese Bulletin of Botany, 2023, 58(6): 905-916. |
| [4] | Na Wang, Teng Jiang, Binxi Wang, Lifang Niu, Hao Lin. Advances in Haploid Breeding Technology and Its Application in Alfalfa and Other Legume Forages [J]. Chinese Bulletin of Botany, 2022, 57(6): 756-763. |
| [5] | He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng. Advance in Gene-editing Technology Based on CRISPR/Cas9 and Its Application in Plants [J]. Chinese Bulletin of Botany, 2022, 57(4): 508-531. |
| [6] | Lubin Tan, Chuanqing Sun. Rapid Domestication of Wild Allotetraploid Rice: Starting a New Era of Human Agricultural Civilization [J]. Chinese Bulletin of Botany, 2021, 56(2): 134-137. |
| [7] | Huijin Fan, Kangming Jin, Renying Zhuo, Guirong Qiao. Cloning and Expression Analysis of Different Truncated U3 Promoters in Phyllostachys edulis [J]. Chinese Bulletin of Botany, 2020, 55(3): 299-307. |
| [8] | Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang. Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing [J]. Chinese Bulletin of Botany, 2019, 54(3): 385-395. |
| [9] | Wang Ying, Li Xianggan, Qiu Lijuan. Research Progress in Off-target in CRISPR/Cas9 Genome Editing [J]. Chinese Bulletin of Botany, 2018, 53(4): 528-541. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||