

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (1): 42-50.DOI: 10.11983/CBB17058 cstr: 32102.14.CBB17058
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Cheng Chen1,2, Aiwu Dong1,2, Wei Su1,2,*( )
)
Received:2017-03-22
Accepted:2017-08-30
Online:2018-01-01
Published:2018-08-10
Contact:
Wei Su
Cheng Chen, Aiwu Dong, Wei Su. Histone Chaperone AtHIRA is Involved in Somatic Homologous Recombination and Salinity Response in Arabidopsis[J]. Chinese Bulletin of Botany, 2018, 53(1): 42-50.
| Primer name | Primer sequence (5′-3′) |
|---|---|
| ACTIN2-F | GGCGATGAAGCTCAATCCAAA |
| ACTIN2-R | GGTCACGACCAGCAAGATCAAG |
| GUS-F | AAGTGGATTGATGTGATATCTC |
| GUS-R | TTCGCGCTGATACCAGACG |
| ATM-F | TGCAGCTGCGTCTCTGCATGA |
| ATM-R | CTTCATGCCGCCCTTGGGCA |
| BRCA1-F | TGCTCAGGGCTCACAGTTGAAGA |
| BRCA1-R | TGCAGGCTCCGTTTTCATTGATTG |
| PARP1-F | TGCTCGCGCGAACTCACTTCT |
| PARP1-R | AGCCTCTCCACCAGAACGGCT |
| PARP2-F | AGCCTGAAGGCCCGGGTAACA |
| PARP2-R | GCTGTCTCAGTTTTGGCTGCCG |
| RAD51-F | CGCCATTTCCCTCCACTCTCAAGC |
| RAD51-R | ACCTGCTGCCTGAAGCTGTTCG |
| RAD54-F | TGAGAGACAGGTGGGCACTCC |
| RAD54-R | ACGTCACCTCGTCACCTGCTGA |
Table 1 Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| ACTIN2-F | GGCGATGAAGCTCAATCCAAA |
| ACTIN2-R | GGTCACGACCAGCAAGATCAAG |
| GUS-F | AAGTGGATTGATGTGATATCTC |
| GUS-R | TTCGCGCTGATACCAGACG |
| ATM-F | TGCAGCTGCGTCTCTGCATGA |
| ATM-R | CTTCATGCCGCCCTTGGGCA |
| BRCA1-F | TGCTCAGGGCTCACAGTTGAAGA |
| BRCA1-R | TGCAGGCTCCGTTTTCATTGATTG |
| PARP1-F | TGCTCGCGCGAACTCACTTCT |
| PARP1-R | AGCCTCTCCACCAGAACGGCT |
| PARP2-F | AGCCTGAAGGCCCGGGTAACA |
| PARP2-R | GCTGTCTCAGTTTTGGCTGCCG |
| RAD51-F | CGCCATTTCCCTCCACTCTCAAGC |
| RAD51-R | ACCTGCTGCCTGAAGCTGTTCG |
| RAD54-F | TGAGAGACAGGTGGGCACTCC |
| RAD54-R | ACGTCACCTCGTCACCTGCTGA |
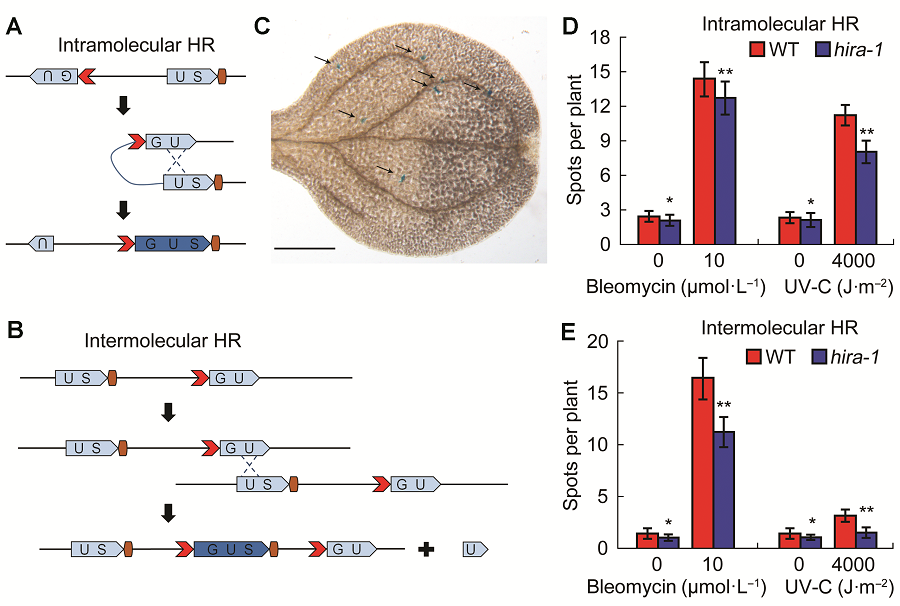
Figure 1 hira1 mutant shows reduced homologus recombination frequency compared with Col-0 of Arabidopsis(A) Scheme of homologus recombination (HR) event in intramolecular line 1445. The two fragments of GUS gene can recombine to form a function GUS gene after a HR event; (B) Scheme of HR event in intermolecular line IC9C. The recombination of separated GUS fragments require intermolecular interaction to restore a functional GUS gene after a HR event; (C) Arabidopsis leaf with arrow labeled blue spots/sectors represent a functional GUS gene, which indicate an independent HR event (Bar=500 μm); (D) Comparison of intramolecular recombination frequency between Col-0 and hira-1; (E) Comparison of intermolecular recombination frequency between Col-0 and hira-1. WT: Wild type; HR: Homologus recombination. * P<0.05; ** P<0.01
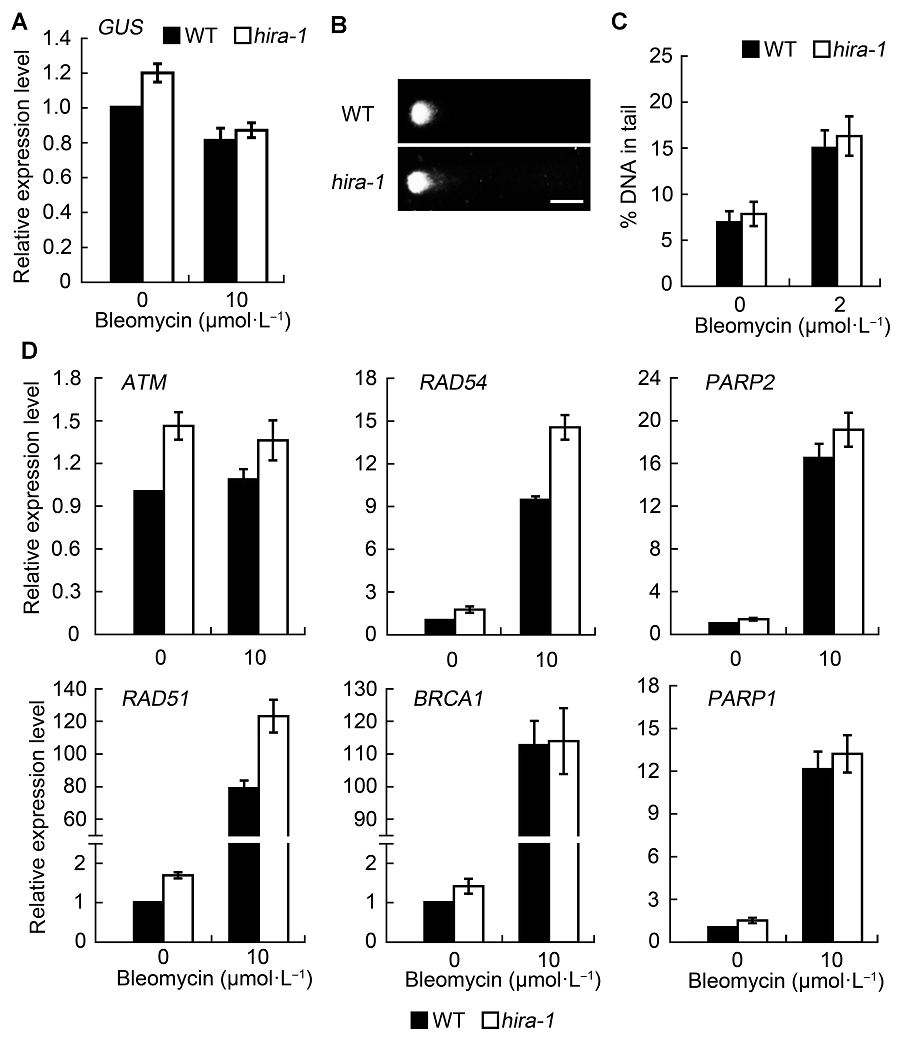
Figure 2 Effect of AtHIRA mutation on DNA damage level and expression level of DNA repair genes (means±SD)(A) Expression level of GUS gene in wild type and hira-1 mutant of Arabidopsis under normal conditions and bleomycin treatment; (B) Representative comet images of wild-type and hira-1 nuclei after bleomycin treatment (Bar=10 μm); (C) The average percentage of DNA in comet tails of wild type and hira-1 mutant. More than 100 individual nuclei were recorded and calculated; (D) Relative expression level of DNA repair genes by RT-qPCR between wild type and hira-1. Three biological repeats were analyzed.
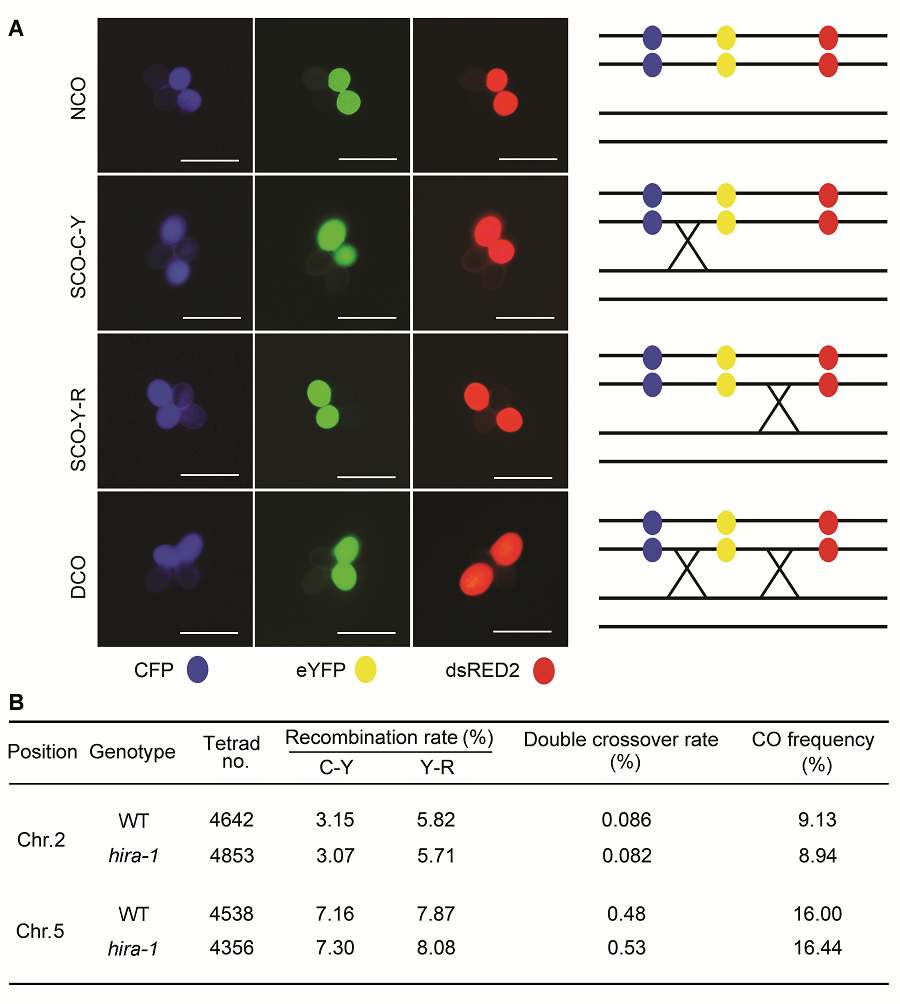
Figure 3 Comparison of meiotic recombination frequency between wild type (WT) and hira-1 mutant of Arabidopsis using the fluorescent tagged line tetrad analysis system(A) Examples of tetrad fluorescent patterns including no cross over (NCO), two types of single cross overs (SCO-C-Y and SCO- Y-R) and one type of double cross over (DCO). The schematic representation of corresponding CO events is shown at right of each tetrad class (Bars=10 μm); (B) Number of each tetrad fluorescent patterns observed in wild type and hira-1.
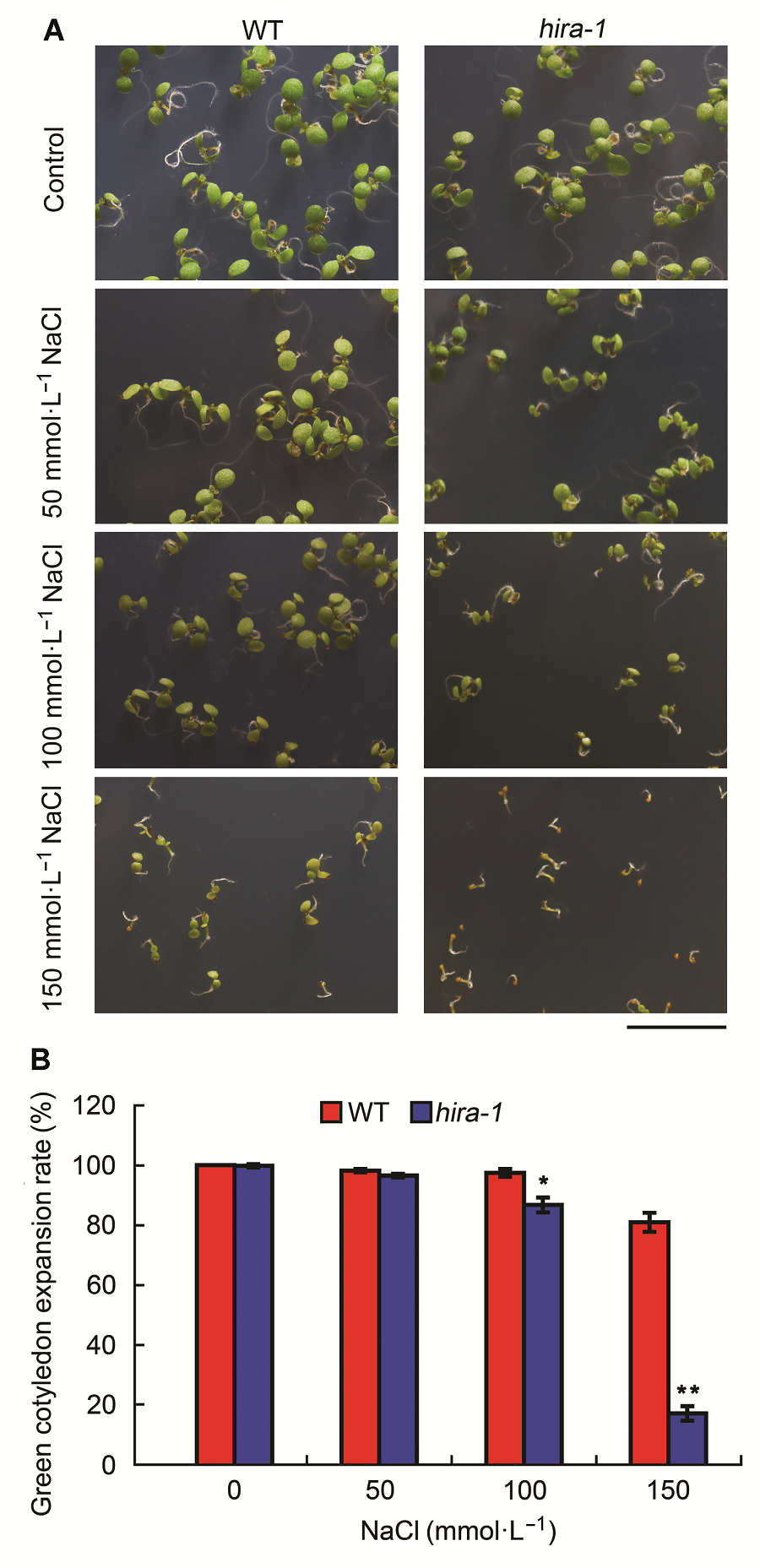
Figure 4 Arabidopsis mutant hira-1 exhibited hypersensitivi- ty to salt stress (means±SD)(A) Phenotypes of 7-day-old seedlings of wild type (WT) and hira-1 grown on media containing 0, 50, 100, and 150 mmol·L-1 NaCl (Bar=1 cm); (B) Comparison of green cotyledon expansion rates of 7-day-old seedlings between wild type and hira-1. Over 100 seeds each genotype were recorded. Three biological repeats were analyzed.
| [1] | 夏志强, 何奕昆, 鲍时来, 种康 (2007). 植物开花的组蛋白甲基化调控分子机理. 植物学通报 24, 275-283. |
| [2] | Amin AD, Vishnoi N, Prochasson P (2012). A global requirement for the HIR complex in the assembly of chromatin.Biochim Biophys Acta 1819, 264-276. |
| [3] | Andersen SL, Sekelsky J (2010). Meiotic versus mitotic recombination: two different routes for double-strand b- reak repair: the different functions of meiotic versus mi- totic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32, 1058-1066. |
| [4] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP (2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [5] | Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell 16, 479-485. |
| [6] | Gao J, Zhu Y, Zhou WB, Molinier J, Dong AW, Shen WH (2012). NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis.Plant Cell 24, 1437-1447. |
| [7] | Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI (2001). Homologous recombination in planta is stimulated in the absence of Rad50.EMBO Rep 2, 287-291. |
| [8] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [9] | Heyer WD, Ehmsen KT, Liu J (2010). Regulation of homologous recombination in eukaryotes.Annu Rev Genet 44, 113-139. |
| [10] | Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010). Zygotic resetting of the HISTONE 3 variant re- pertoire participates in epigenetic reprogramming in Arabi- dopsis.Curr Biol 20, 2137-2143. |
| [11] | Krejci L, Altmannova V, Spirek M, Zhao XL (2012). Homologous recombination and its regulation.Nucleic Acids Res 40, 5795-5818. |
| [12] | Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P (2005). The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus.Nature 437, 1386-1390. |
| [13] | Melamed-Bessudo C, Levy AA (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchro- matic but not in heterochromatic regions in Arabidopsis.Proc Natl Acad Sci USA 109, E981-E988. |
| [14] | Molinier J, Ries G, Bonhoeffer S, Hohn B (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16, 342-352. |
| [15] | Nie X, Wang HF, Li J, Holec S, Berger F (2014). The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics.Biol Open 3, 794-802. |
| [16] | Okada T, Endo M, Singh MB, Bhalla PL (2005). Analysis of the histoneH3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44, 557-568. |
| [17] | Otero S, Desvoyes B, Gutierrez C (2014). Histone H3 dynamics in plant cell cycle and development.Cytogenet Genome Res 143, 114-124. |
| [18] | Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ (2002). Targeted mutagenesis of theHira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryo- nic lethality. Mol Cell Biol 22, 2318-2328. |
| [19] | Schuermann D, Fritsch O, Lucht JM, Hohn B (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase δ mutants.Plant Cell 21, 2700-2714. |
| [20] | Schuermann D, Molinier J, Fritsch O, Hohn B (2005). The dual nature of homologous recombination in plants.Tren- ds Genet 21, 172-181. |
| [21] | Sherwood PW, Osley MA (1991). Histone regulatory (hir) mutations suppress δ insertion alleles in Saccharomyces cerevisiae. Genetics 128, 729-738. |
| [22] | Szenker E, Lacoste N, Almouzni G (2012). A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep 1, 730-740. |
| [23] | Tang Y, Poustovoitov MV, Zhao KH, Garfinkel M, Canu- tescu A, Dunbrack R, Adams PD, Marmorstein R (2006). Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly.Nat Struct Mol Biol 13, 921-929. |
| [24] | Wijeratne AJ, Ma H (2007). Genetic analyses of meiotic recombination in Arabidopsis. J Integr Plant Biol 49, 1199-1207. |
| [25] | Xu Y, Price BD (2011). Chromatin dynamics and the repair of DNA double strand breaks.Cell Cycle 10, 261-267. |
| [26] | Zhu Y, Dong AW, Meyer D, Pichon O, Renou JP, Cao KW, Shen WH (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18, 2879-2892. |
| [1] | Xu Tiantian, Yang Peijian, Zhou Xiaoxi, Cao Yi, Chen Yanhong, Liu Guoyuan, Zhang Jian, Wei Hui. Analysis of Physicochemical Characteristics and Expression Characteristics of Lagerstroemia indica GolS Family Genes [J]. Chinese Bulletin of Botany, 2025, 60(3): 393-406. |
| [2] | Jinyu Du, Zhen Sun, Yanlong Su, Heping Wang, Yaling Liu, Zhenying Wu, Feng He, Yan Zhao, Chunxiang Fu. Identification and Functional Analysis of an Agropyron mongolicum Caffeic Acid 3-O-methyltransferase Gene AmCOMT1 [J]. Chinese Bulletin of Botany, 2024, 59(3): 383-396. |
| [3] | Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149. |
| [4] | Qi Zhang, Wenjing Zhang, Xiankai Yuan, Ming Li, Qiang Zhao, Yanli Du, Jidao Du. The Regulatory Mechanism of Melatonin on Nucleic Acid Repairing of Common Bean (Phaseolus vulgaris) at the Sprout Stage Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 108-121. |
| [5] | Nan Zhang,Ziguang Liu,Shichen Sun,Shengyi Liu,Jianhui Lin,Yifang Peng,Xiaoxu Zhang,He Yang,Xi Cen,Juan Wu. Response of AtR8 lncRNA to Salt Stress and Its Regulation on Seed Germination in Arabidopsis [J]. Chinese Bulletin of Botany, 2020, 55(4): 421-429. |
| [6] | Dongdong Cao,Shanyu Chen,Yebo Qin,Huaping Wu,Guanhai Ruan,Yutao Huang. Regulatory Mechanism of Salicylic Acid on Seed Germination Under Salt Stress in Kale [J]. Chinese Bulletin of Botany, 2020, 55(1): 49-61. |
| [7] | Lulu Li,Wenchao Yin,Mei Niu,Wenjing Meng,Xiaoxing Zhang,Hongning Tong. Functional Analysis of Brassinosteroids in Salt Stress Responses in Rice [J]. Chinese Bulletin of Botany, 2019, 54(2): 185-193. |
| [8] | Chen Xu, Xiaolong Liu, Qian Li, Fenglou Ling, Zhihai Wu, Zhian Zhang. Effect of Salt Stress on Photosynthesis and Chlorophyll Fluorescence Characteristics of Rice Leaf for Nitrogen Levels [J]. Chinese Bulletin of Botany, 2018, 53(2): 185-195. |
| [9] | Shuhua Guo, Yongjiang Sun, Yanjie Niu, Ning Han, Heng Zhai, Yuanpeng Du. Effect of Alkaline Salt Stress on Photosystem Activity of Grape F1 Generation Hybrids [J]. Chinese Bulletin of Botany, 2018, 53(2): 196-202. |
| [10] | Baoling Liu, Li Zhang, Yan Sun,Jinai Xue, Changyong Gao, Lixia Yuan, Jiping Wang, Xiaoyun Jia, Runzhi Li. Genome-wide Characterization of bZIP Transcription Factors in Foxtail Millet and Their Expression Profiles in Response to Drought and Salt Stresses [J]. Chinese Bulletin of Botany, 2016, 51(4): 473-487. |
| [11] | Qing-Xian KONG, Jiang-Bao XIA, Zi-Guo ZHAO, Fan-Zhu QU. Effects of groundwater salinity on the characteristics of leaf photosynthesis and stem sap flow in Tamarix chinensis [J]. Chin J Plan Ecolo, 2016, 40(12): 1298-1309. |
| [12] | Lin Qi, Xinfu Bai, Weihao Niu, Zhenhua Zhang. Effect of Rhizosphere Ventilation on Growth of Cotton Seedlings Under Salt Stress [J]. Chinese Bulletin of Botany, 2016, 51(1): 16-23. |
| [13] | GUO Rui,LI Feng,ZHOU Ji,LI Hao-Ru,XIA Xu,LIU Qi. Eco-physiological responses of linseed (Linum usitatissimum) to salt and alkali stresses [J]. Chin J Plan Ecolo, 2016, 40(1): 69-79. |
| [14] | Qiong Jiang, Youning Wang, Lixiang Wang, Zhengxi Sun, Xia Li. Validation of Reference Genes for Quantitative RT-PCR Analysis in Soybean Root Tissue under Salt Stress [J]. Chinese Bulletin of Botany, 2015, 50(6): 754-764. |
| [15] | MENG De-Yun,HOU Lin-Lin,YANG Sha,MENG Jing-Jing,GUO Feng,LI Xin-Guo,WAN Shu-Bo. Exogenous polyamines alleviating salt stress on peanuts (Arachis hypogaea) grown in pots [J]. Chin J Plan Ecolo, 2015, 39(12): 1209-1215. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||