

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (2): 175-183.DOI: 10.11983/CBB15054 cstr: 32102.14.CBB15054
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Jiaxiao Du, Zongyan Qin, Si Xu, Xiang Jing, Ying Bao*( )
)
Received:2015-03-31
Accepted:2015-09-16
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: Jiaxiao Du, Zongyan Qin, Si Xu, Xiang Jing, Ying Bao. Phylogenetic Origin of BBCC Genome Allotetraploids in Oryza Revealed by Chloroplast Gene Sequences[J]. Chinese Bulletin of Botany, 2016, 51(2): 175-183.
| Species | Genome | IRRI Acc. No. | Origin |
|---|---|---|---|
| Oryza punctata Kotechy ex Steud. | BB | 103888 | Tanzania |
| 101434 | Tanzania | ||
| O. eichingeri A. Peter | CC | 105159 | Uganda |
| O. officinalis Wall. ex G. Watt. | CC | 104973 | China |
| O. rhizomatis D. A. Vaughan | CC | 103410 | Sri Lanka |
| O. malampuzhaensis Krish. et Chand. | BBCC | 80764 | India |
| 105223 | India | ||
| 105328 | India | ||
| O. minuta J. S. Presl. ex C. B. Presl. | BBCC | 104677 | Philippines |
| 105307 | Philippines | ||
| 103880 | Philippines | ||
| O. punctata | BBCC | 105181 | Uganda |
| 104059 | Nigeria | ||
| 104975 | Kenya | ||
| O. australiensis Domin | EE | 103303 | Australia |
| 105165 | Australia |
Table 1 Plant materials of Oryza used in this study
| Species | Genome | IRRI Acc. No. | Origin |
|---|---|---|---|
| Oryza punctata Kotechy ex Steud. | BB | 103888 | Tanzania |
| 101434 | Tanzania | ||
| O. eichingeri A. Peter | CC | 105159 | Uganda |
| O. officinalis Wall. ex G. Watt. | CC | 104973 | China |
| O. rhizomatis D. A. Vaughan | CC | 103410 | Sri Lanka |
| O. malampuzhaensis Krish. et Chand. | BBCC | 80764 | India |
| 105223 | India | ||
| 105328 | India | ||
| O. minuta J. S. Presl. ex C. B. Presl. | BBCC | 104677 | Philippines |
| 105307 | Philippines | ||
| 103880 | Philippines | ||
| O. punctata | BBCC | 105181 | Uganda |
| 104059 | Nigeria | ||
| 104975 | Kenya | ||
| O. australiensis Domin | EE | 103303 | Australia |
| 105165 | Australia |
| Gene | Primer sequences (5'-3') |
|---|---|
| atpA | AGGCTTACTTGGGTCGTGTT TCCTCGGTGAATGTCTTGCT |
| atpB | ATCCTACTACTTCTCGTCCC AATTGCGATAATGTCCTGAA |
| petA | TTGGGTAAAGGAACAGATGA AATAACGGATGCGAAGAAGA |
| psaA | TAGCAGGGTTATTAGGACTTG ATGCGTGAATGTGATGGACT |
| psbA | TCGGATGGTTCGGTGTTTTGA AGGGAAGTTGTGAGCATTACG |
| psbB | GGGTTTGCCTTGGTATCGTG GGCTGTCTCCCTGTAGTTGG |
| psbC | GGGTTGATTTTACTTCCGCAC GAAGGTCCCAAAAACGCATAG |
| rbcL | TATCTTGGCAGCATTCCGAGTAA ATTTGATCGCTTTCCATATTTC |
Table 2 The sequences of primers
| Gene | Primer sequences (5'-3') |
|---|---|
| atpA | AGGCTTACTTGGGTCGTGTT TCCTCGGTGAATGTCTTGCT |
| atpB | ATCCTACTACTTCTCGTCCC AATTGCGATAATGTCCTGAA |
| petA | TTGGGTAAAGGAACAGATGA AATAACGGATGCGAAGAAGA |
| psaA | TAGCAGGGTTATTAGGACTTG ATGCGTGAATGTGATGGACT |
| psbA | TCGGATGGTTCGGTGTTTTGA AGGGAAGTTGTGAGCATTACG |
| psbB | GGGTTTGCCTTGGTATCGTG GGCTGTCTCCCTGTAGTTGG |
| psbC | GGGTTGATTTTACTTCCGCAC GAAGGTCCCAAAAACGCATAG |
| rbcL | TATCTTGGCAGCATTCCGAGTAA ATTTGATCGCTTTCCATATTTC |
| Gene name | Length (bp) | SNP | GC (%) |
|---|---|---|---|
| atpA | 997 | 3 | 42 |
| atpB | 673 | 3 | 43 |
| matK | 1552 | 16 | 34 |
| petA | 770 | 0 | 43 |
| psaA | 511 | 1 | 44 |
| psbA | 662 | 3 | 42 |
| psbB | 628 | 1 | 45 |
| psbC | 648 | 5 | 41 |
| rbcL | 1019 | 6 | 43 |
| Total | 7460 | 41 | 42 |
Table 3 Sequence characteristics for 9 chloroplast gene fragments of Oryza
| Gene name | Length (bp) | SNP | GC (%) |
|---|---|---|---|
| atpA | 997 | 3 | 42 |
| atpB | 673 | 3 | 43 |
| matK | 1552 | 16 | 34 |
| petA | 770 | 0 | 43 |
| psaA | 511 | 1 | 44 |
| psbA | 662 | 3 | 42 |
| psbB | 628 | 1 | 45 |
| psbC | 648 | 5 | 41 |
| rbcL | 1019 | 6 | 43 |
| Total | 7460 | 41 | 42 |
| Species (genome) | Variable nucleotide sites | |||||||
|---|---|---|---|---|---|---|---|---|
| atpA | atpB | matK | psaA | psbA | psbB | psbC | rbcL | |
| O. punctata (BB) | GGA | GGC | CTATATAATTCTTTCT | C | ACC | G | GAAA | CTTGTC |
| O. punctata (BB) | ... | ... | ................ | . | ... | . | .... | ...... |
| O. minuta (BBCC) | A.. | ... | ................ | . | ... | . | .... | ...... |
| O. malampuzhaensis (BBCC) | A.. | A.. | .......C........ | . | ... | . | .... | ...... |
| O. eichingeri (CC) | AAC | AAT | TAGCGCCATCAACTTA | T | GTT | A | GGCG | ACGACG |
| O. officinalis (CC) | ... | ... | ........A....C.. | . | ... | . | C.A. | ...... |
| O. rhizomatis (CC) | ... | ... | ................ | . | ... | . | ..A. | ...... |
| O. punctata (BBCC) | ... | ... | ................ | . | ... | . | .... | ...... |
Table 4 Variable nucleotide sites of 9 chloroplast gene fragments of Oryza
| Species (genome) | Variable nucleotide sites | |||||||
|---|---|---|---|---|---|---|---|---|
| atpA | atpB | matK | psaA | psbA | psbB | psbC | rbcL | |
| O. punctata (BB) | GGA | GGC | CTATATAATTCTTTCT | C | ACC | G | GAAA | CTTGTC |
| O. punctata (BB) | ... | ... | ................ | . | ... | . | .... | ...... |
| O. minuta (BBCC) | A.. | ... | ................ | . | ... | . | .... | ...... |
| O. malampuzhaensis (BBCC) | A.. | A.. | .......C........ | . | ... | . | .... | ...... |
| O. eichingeri (CC) | AAC | AAT | TAGCGCCATCAACTTA | T | GTT | A | GGCG | ACGACG |
| O. officinalis (CC) | ... | ... | ........A....C.. | . | ... | . | C.A. | ...... |
| O. rhizomatis (CC) | ... | ... | ................ | . | ... | . | ..A. | ...... |
| O. punctata (BBCC) | ... | ... | ................ | . | ... | . | .... | ...... |
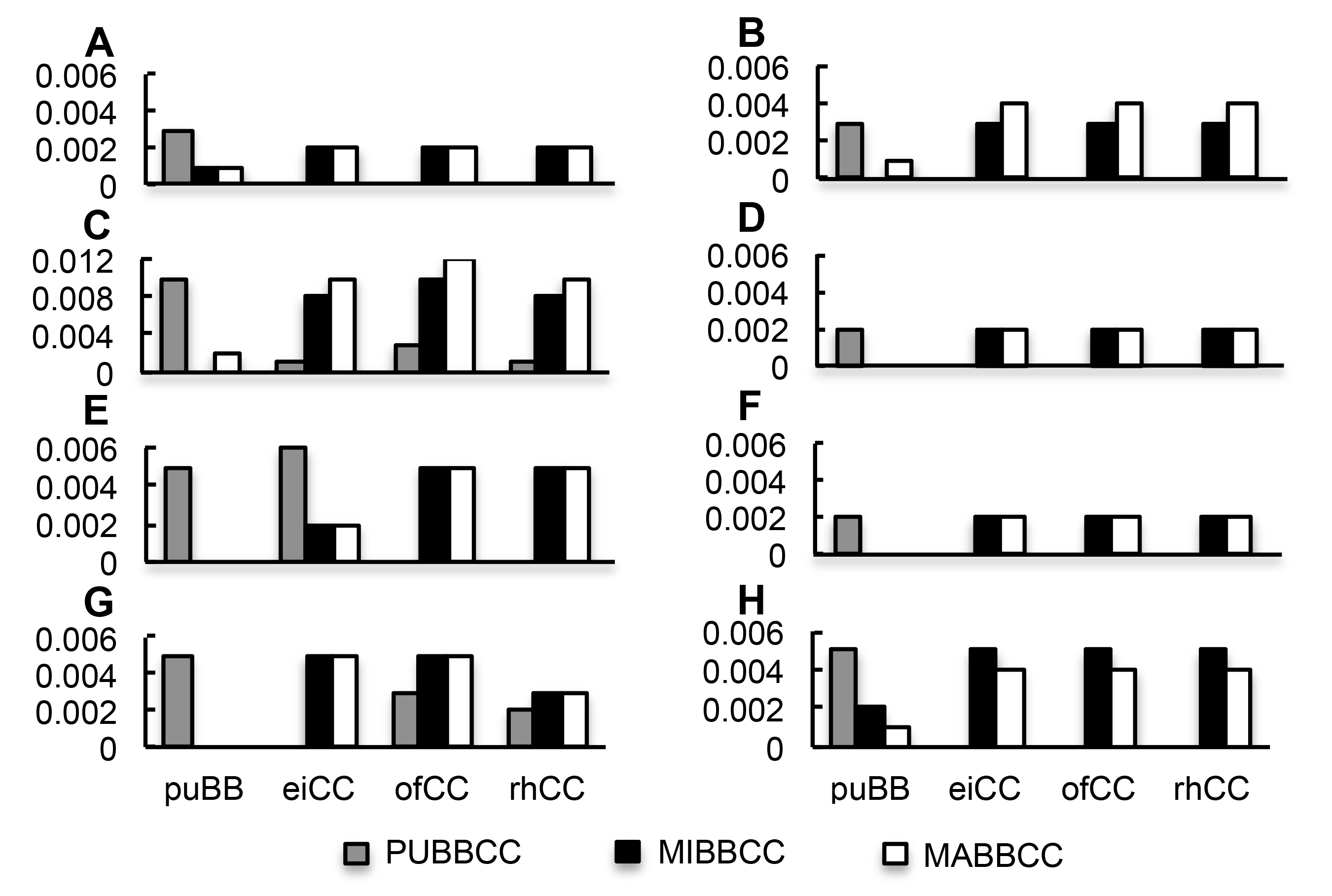
Figure 1 Pairwise genetic distances of the BBCC genomic tetraploids and their related diploids (A)-(H) Pairwise genetic distances of atpA, atpB, matK, psaA, psbA, psbB, psbC, and rbcL, respectively. x axis: puBB, eiCC, ofCC, and rhCC represents diploid O. punctata with genome BB, and O. eichingeri, O. officinalis, and O. rhizomatis with genome CC, respectively; y axis: Genetic distances

Figure 2 The maximum likelihood trees based on 8 and combined chloroplast genes (A)-(H) The ML tree based on atpA, atpB, matK, psaA, psbA, psbB, psbC, and rbcL gene, respectively; (I) The ML tree based on 5 combined genes (atpA, atpB, matK, psbA, and psbC). Numbers near branches indicate bootstrap values above 50%, Boldface indicates allotetraploid species with BBCC genome.
| [1] | Aggarwal RK, Brar DS, Khush GS (1997). Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254, 1-12. |
| [2] | Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999). Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98, 1320-1328. |
| [3] | Avise JC (2000). Phylogeography:the History and Forma- tion of Species. Cambridge: Harvard University Press. pp. 1-447. |
| [4] | Bao Y, Ge S (2003). Phylogenetic relationships among diploid species of Oryza officinalis complex revealed by multiple gene sequences. Acta Phytot Sin 41, 497-508. |
| [5] | Bao Y, Lu B, Ge S (2005). Identification of genomic consti- tutions of Oryza species with the B and C genomes by the PCR-RFLP method. Genet Res Crop Evol 52, 69-76. |
| [6] | Bao Y, Wendel FJ, Ge S (2010). Multiple patterns of rDNA evolution following polyploidy in Oryza. Mol Phylogenet Evol 55, 136-142. |
| [7] | Bao Y, Zhou HF, Ge S, Hong DY (2006). Genetic diversity and evolutionary relationships of Oryza species with the B- and C-genomes as revealed by SSR markers. J Plant Biol 49, 339-347. |
| [8] |
Buso GS, Rangel PH, Ferreira ME (2001). Analysis of random and specific sequences of nuclear and cytoplas- mic DNA in diploid and tetraploid American wild rice species (Oryza spp.). Genome 44, 476-494.
DOI PMID |
| [9] |
Criscuolo A (2011). morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol 61, 944-948.
DOI PMID |
| [10] |
Dally AM, Second G (1990). Chloroplast DNA diversity in wild and cultivated species of rice (genus Oryza, section Oryza). Cladistic-mutation and genetic-distance analysis. Theor Appl Genet 80, 209-222.
DOI PMID |
| [11] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [12] | Ge S, Li A, Lu BR, Zhang SZ, Hong DY (2002). A phylogeny of the rice tribe Oryzeae (Poaceae) based on matK sequence data. Am J Bot 89, 1967-1972. |
| [13] |
Ge S, Sang T, Lu BR, Hong DY (1999). Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96, 14400-14405.
DOI PMID |
| [14] |
Ge S, Sang T, Lu BR, Hong DY (2001). Rapid and reliable identification of rice genomes by RFLP analysis of PCR- amplified Adh genes. Genome 44, 1136-1142.
PMID |
| [15] |
Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27, 221-224.
DOI PMID |
| [16] |
Khush GS (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35, 25-34.
PMID |
| [17] | Kumagai M, Wang L, Ueda S (2010). Genetic diversity and evolutionary relationships in genus Oryza revealed by using highly variable regions of chloroplast DNA. Gene 462, 44-51. |
| [18] | Lu BR, Ge S, Sang T, Chen JK, Hong DY (2001). The current taxonomy and perplexity of the genus Oryza (Poaceae). Acta Phytot Sin 39, 373-388. |
| [19] |
Nishikawa T, Vaughan DA, Kadowaki K (2005). Phyloge- netic analysis of Oryza species, based on simple se- quence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theor Appl Genet 110, 696-705.
PMID |
| [20] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731-2739.
DOI PMID |
| [21] | Tian XJ, Zheng J, Hu SN, Yu J (2006). The rice mitochon- drial genomes and their variations. Plant Physiol 140, 401-410. |
| [22] | Vaughan DA (1994). The Wild Relatives of Rice:a Genetic Resources Handbook. Manila: International Rice Rese- arch Institute. pp. 1-137. |
| [23] | Wang BS, Ding ZY, Liu W, Pan J, Li CB, Ge S, Zhang DM (2009). Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol Biol 9, 250. |
| [24] |
Wing RA, Ammiraju JS, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, Brar D, Mackill DJ, Han B, Soderlund C, Stein L, SanMiguel P, Jackson S (2005). The Oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol 59, 53-62.
DOI PMID |
| [1] | Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome and Phylogenetic Inference of the Aronia melanocarpa [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [3] | Zhang Ruli, Li Dezhu, Zhang Yuxiao. Population Genetic Structure and Climate Adaptation Analysis of Brachystachyum densiflorum [J]. Chinese Bulletin of Botany, 2025, 60(3): 407-424. |
| [4] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [5] | Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng. Progresses in the study of the relationship between plant genome size and traits [J]. Biodiv Sci, 2025, 33(2): 24192-. |
| [6] | Linfeng Xia, Rui Li, Haizheng Wang, Daling Feng, Chunyang Wang. Research Advances and Prospects in Charophytes Genomics [J]. Chinese Bulletin of Botany, 2025, 60(2): 271-282. |
| [7] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [8] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [9] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [10] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| [11] | Danling Hu, Yongwei Sun. Advances in Virus-mediated Genome Editing Technology in Plants [J]. Chinese Bulletin of Botany, 2024, 59(3): 452-462. |
| [12] | Zhi Yang, Yong Yang. Research Advances on Nuclear Genomes of Economically Important Trees of Lauraceae [J]. Chinese Bulletin of Botany, 2024, 59(2): 302-318. |
| [13] | Zhengyong Duan, Min Ding, Yuzhuo Wang, Yibing Ding, Ling Chen, Ruiyun Wang, Zhijun Qiao. Genome-wide Identification and Expression Analysis of SBP Genes in Panicum miliaceum [J]. Chinese Bulletin of Botany, 2024, 59(2): 231-244. |
| [14] | Xiting Yu, Xuehui Huang. New Insights Into the Origin of Modern Maize-hybridization of Two Teosintes [J]. Chinese Bulletin of Botany, 2023, 58(6): 857-860. |
| [15] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||