

植物学报 ›› 2020, Vol. 55 ›› Issue (3): 299-307.DOI: 10.11983/CBB19217 cstr: 32102.14.CBB19217
收稿日期:2019-11-07
接受日期:2020-03-24
出版日期:2020-05-01
发布日期:2020-07-06
通讯作者:
乔桂荣
基金资助:
Huijin Fan1,2,Kangming Jin1,Renying Zhuo1,Guirong Qiao1,*( )
)
Received:2019-11-07
Accepted:2020-03-24
Online:2020-05-01
Published:2020-07-06
Contact:
Guirong Qiao
摘要: 具有明确转录起始位点的U3和U6启动子是CRISPR/Cas9技术中驱动sgRNA转录的重要元件。从毛竹(Phyllostachys edulis)中克隆了2个PeU3启动子, 均进行了3个不同长度的截短, 长度分别为550、397和149 bp及561、392和152 bp; 并分别构建6个启动子驱动的GUS及LUC植物表达载体, 再利用农杆菌(Agrobacterium tumefaciens)介导法分别转化麻竹(Dendrocalamus latiflorus)愈伤组织和烟草(Nicotiana benthamiana)叶片。结果显示, 这些PeU3启动子总体都具有转录活性, 不同PeU3启动子以及同一PeU3启动子不同截短时其转录活性不同, 其中长度为397 bp的PeU3-1-2pro启动子活性最强, 可为构建竹子CRISPR/Cas9基因组编辑体系提供更多理想的内源启动子。
凡惠金, 金康鸣, 卓仁英, 乔桂荣. 毛竹不同截短U3启动子的克隆及表达分析. 植物学报, 2020, 55(3): 299-307.
Huijin Fan, Kangming Jin, Renying Zhuo, Guirong Qiao. Cloning and Expression Analysis of Different Truncated U3 Promoters in Phyllostachys edulis. Chinese Bulletin of Botany, 2020, 55(3): 299-307.
| Primer name | Sequences (5'-3') |
|---|---|
| PeU3-1 | F: ATCCGCCCCTGGTTGCTCTA R: CAAGCCGTTAATCACGCTCTGG |
| PeU3-2 | F: TCAGCGTTGACCTCCTCTTG R: TGGCACGAAATGGGATGAAG |
| 1301-PeU3-1-1-F | F: GACCTGCAGGCATGCAAGCTTTCAGCGTTGACCTCCTCTT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-2-F | F: GACCTGCAGGCATGCAAGCTCAGGTCATAAGATGGAAGCT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-3-F | F: GACCTGCAGGCATGCAAGCTCAGCCCATACGAAAGTGATT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-2-1-F | F: GACCTGCAGGCATGCAAGCTCCCTAGTGATGCCTTAAAGC R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-2-F | F: GACCTGCAGGCATGCAAGCTATGGTATTCTGTTGCGGACT R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-3-F | F: GACCTGCAGGCATGCAAGCTTGGCCTTTTTCATGGAGCCG R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-1-1-F | F: TCGACGGTATCGATAAGCTTTTCAGCGTTGACCTCCTCTT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-2-F | F: TCGACGGTATCGATAAGCTTCAGGTCATAAGATGGAAGCT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-3-F | F: TCGACGGTATCGATAAGCTTCAGCCCATACGAAAGTGATT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-2-1-F | F: TCGACGGTATCGATAAGCTTATCCGCCCCTGGTTGCTCTA R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-2-F | F: TCGACGGTATCGATAAGCTTATGGTATTCTGTTGCGGACT R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-3-F | F: TCGACGGTATCGATAAGCTTTGGCCTTTTTCATGGAGCCG R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
表1 本研究中所使用的引物序列
Table 1 Primer sequences used in this study
| Primer name | Sequences (5'-3') |
|---|---|
| PeU3-1 | F: ATCCGCCCCTGGTTGCTCTA R: CAAGCCGTTAATCACGCTCTGG |
| PeU3-2 | F: TCAGCGTTGACCTCCTCTTG R: TGGCACGAAATGGGATGAAG |
| 1301-PeU3-1-1-F | F: GACCTGCAGGCATGCAAGCTTTCAGCGTTGACCTCCTCTT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-2-F | F: GACCTGCAGGCATGCAAGCTCAGGTCATAAGATGGAAGCT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-3-F | F: GACCTGCAGGCATGCAAGCTCAGCCCATACGAAAGTGATT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-2-1-F | F: GACCTGCAGGCATGCAAGCTCCCTAGTGATGCCTTAAAGC R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-2-F | F: GACCTGCAGGCATGCAAGCTATGGTATTCTGTTGCGGACT R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-3-F | F: GACCTGCAGGCATGCAAGCTTGGCCTTTTTCATGGAGCCG R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-1-1-F | F: TCGACGGTATCGATAAGCTTTTCAGCGTTGACCTCCTCTT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-2-F | F: TCGACGGTATCGATAAGCTTCAGGTCATAAGATGGAAGCT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-3-F | F: TCGACGGTATCGATAAGCTTCAGCCCATACGAAAGTGATT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-2-1-F | F: TCGACGGTATCGATAAGCTTATCCGCCCCTGGTTGCTCTA R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-2-F | F: TCGACGGTATCGATAAGCTTATGGTATTCTGTTGCGGACT R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-3-F | F: TCGACGGTATCGATAAGCTTTGGCCTTTTTCATGGAGCCG R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
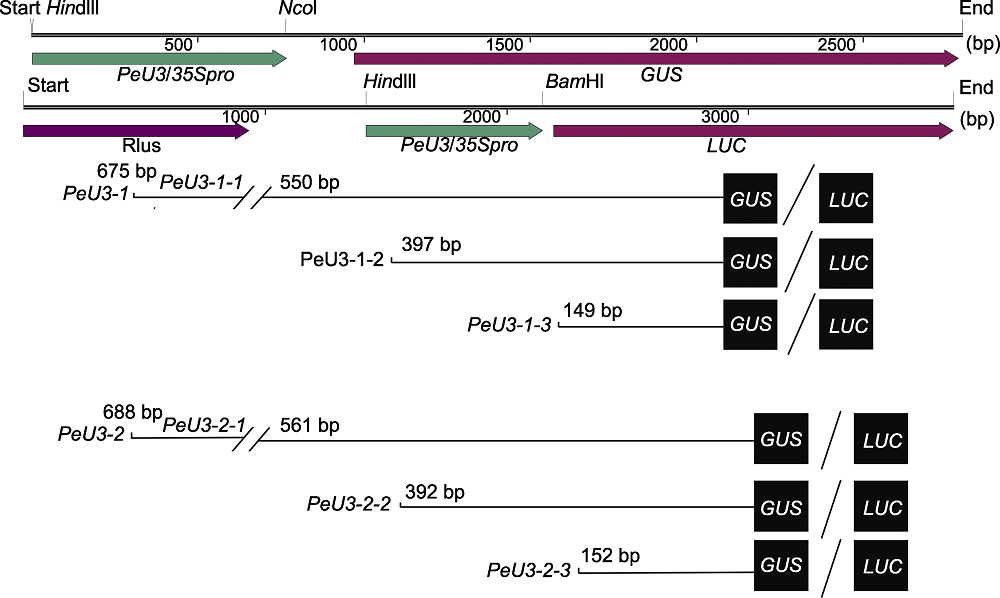
图1 不同截短PeU3启动子驱动报告基因GUS/LUC的表达载体构建
Figure 1 Construction of expression vectors with GUS/LUC reporter gene driven by different truncated PeU3 promoters
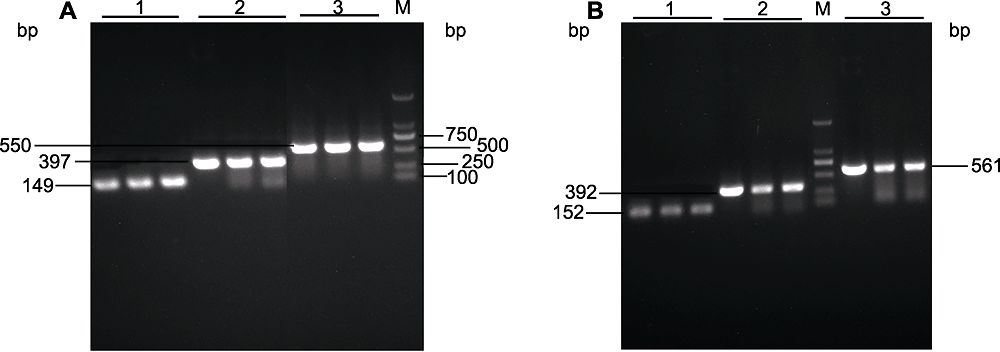
图2 6个不同截短PeU3启动子片段的扩增产物电泳检测 (A) 以PeU3-1启动子质粒为模板进行扩增(1: PeU3-1-3pro; 2: PeU3-1-2pro; 3: PeU3-1-1pro); (B) 以PeU3-2启动子质粒为模板进行扩增(1: PeU3-2-3pro; 2: PeU3-2-2pro; 3: PeU3-2-1pro). M: 2 kb DNA marker
Figure 2 PCR analysis of six truncated PeU3 promoters (A) Amplification using PeU3-1 promoter plasmid as template (1: PeU3-1-3pro; 2: PeU3-1-2pro; 3: PeU3-1-1pro); (B) Amplification using PeU3-2 promoter plasmid as template (1: PeU3-2-3pro; 2: PeU3-2-2pro; 3: PeU3-2-1pro). M: 2 kb DNA marker

图4 不同截短PeU3启动子驱动GUS表达 (A) 不同截短PeU3启动子驱动GUS在烟草叶片中的瞬时表达(Bars=5 mm); (B) 不同截短PeU3启动子驱动GUS在麻竹愈伤组织中的表达(Bars=5 mm)。CK: 阴性对照
Figure 4 GUS expression driven by different truncated PeU3 promoters (A) Transient expression of GUS gene driven by different truncated PeU3 promoters in the leaves of Nicotiana benthamiana (Bars=5 mm); (B) Expression of GUS gene driven by different truncated PeU3 promoters in the callus of Dendrocalamus latiflorus (Bars=5 mm). CK: Negative control
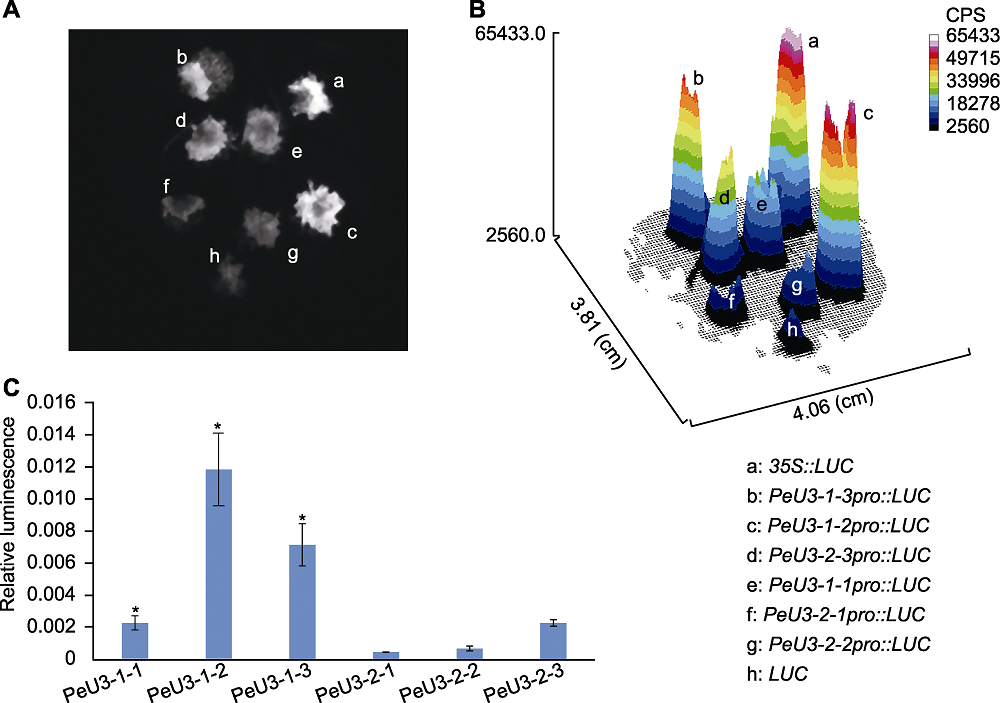
图5 不同截短PeU3启动子驱动LUC在烟草叶片中的瞬时表达 (A), (B) PeU3启动子在烟草叶片中的瞬时表达LUC荧光图; (C) PeU3启动子在烟草叶片中的瞬时表达LUC相对活性(每个实验均设4个生物学重复。* 表示Duncan法检测时在P<0.05水平差异显著); CPS: 荧光强度
Figure 5 LUC expression driven by different truncated PeU3 promoters in the leaves of Nicotiana benthamiana (A), (B) Transient expression LUC assays illustrating the activation of PeU3 promoter in leaves of N. benthamiana; (C) The quantification of the relative luminescence intensities (In each experiment, four biological replications were performed with quanti?cation. Asterisk above the bars denote significant differences determined by the Duncan’s test, P<0.05); CPS: Luminescence intensity
| [1] | 陈香嵩, 李甜甜, 周少立, 赵毓 (2018). 外源蛋白在烟草叶片瞬时表达. BioProtoc doi: 10.217691BioProtoc.1010127. |
| [2] | 雷建峰, 李月, 徐新霞, 阿尔祖古丽·塔什, 蒲艳, 张巨松, 刘晓东 (2016a). 棉花不同GbU6启动子截短克隆及功能鉴定. 作物学报 42, 675-683. |
| [3] | 雷建峰, 徐新霞, 代培红, 李继洋, 张巨松, 刘晓东 (2016b). 不同截短U3启动子在棉花中的功能分析. 棉花学报 28, 307-314. |
| [4] | 李继洋, 雷建峰, 代培红, 姚瑞, 曲延英, 陈全家, 李月, 刘晓东 (2018). 基于棉花U6启动子的海岛棉CRISPR/Cas9基因组编辑体系的建立. 作物学报 44, 227-235. |
| [5] | 李丽莉, 扈廷茂, 扈会平, 刘明秋, 苏慧敏 (2005). 利用番茄U3snRNA基因上游启动区构建植物表达载体及对烟草的转化. 内蒙古大学学报(自然科学版) 36, 63-67. |
| [6] | 蒲艳, 刘晓东, 阿尔祖古丽·塔什, 魏倩, 刘超 (2019). 番茄不同截短U3启动子的克隆及功能分析. 华北农学报 34, 33-39. |
| [7] | 孙建飞, 翟建云, 马元丹, 傅卢成, 卜柯丽, 王柯杨, 高岩, 张汝民 (2018). 毛竹快速生长期茎秆不同节间光合色素和光合酶活性的差异. 植物学报 53, 773-781. |
| [8] | 藏旭阳, 代培红, 李继洋, 蒲艳, 顾爱星, 刘晓东 (2019). 棉花U3和U6启动子在CRISPR/Cas9基因组编辑体系中的功能鉴定. 棉花学报 31, 31-39. |
| [9] | Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug II RG, Tan WF, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114-118. |
| [10] | Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39. |
| [11] |
Bibikova M, Beumer K, Trautman JK, Carroll D (2003). Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764-764.
URL PMID |
| [12] | Bruegmann T, Fladung M (2019). Overexpression of both flowering time genes AtSOC1 and SaFUL revealed huge influence onto plant habitus in poplar. Tree Genet Genomes 15, 20. |
| [13] | Charrier A, Vergen E, Dousset N, Richer A, Petiteau A, Chevreau E (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR- Cas9 system. Front Plant Sci 10, 40. |
| [14] | Chen JR, Shafi M, Li S, Wang J, Wu JS, Ye ZQ, Peng DL, Yan WB, Liu D (2015). Copper induced oxidative stresses, antioxidant responses and phytoremediation potential of Moso bamboo (Phyllostachys pubescens). Sci Rep 5, 13554. |
| [15] | Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. |
| [16] | Doll NM, Gilles LM, Gérentes MF, Richard C, Just J, Fierlej Y, Borrelli VMG, Gendrot G, Ingram GC, Rogowsky PM, Widiez T (2019). Single and multiple gene knockouts by CRISPR-Cas9 in maize. Plant Cell Rep 38, 487-501. |
| [17] |
Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67-71.
DOI URL PMID |
| [18] |
Ge W, Zhang Y, Cheng ZC, Hou D, Li XP, Gao J (2017). Main regulatory pathways, key genes and microRNAs involved in flower formation and development of Moso bamboo (Phyllostachys edulis). Plant Biotechnol J 15, 82-96.
URL PMID |
| [19] |
Jinek M, Chylinski K, Fonfara L, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
URL PMID |
| [20] | Lee K, Zhang YX, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi YP, Wang K (2019). Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17, 362-372. |
| [21] |
Li L, Cheng ZC, Ma YJ, Bai QS, Li XY, Cao ZH, Wu ZN, Gao J (2018). The association of hormone signaling genes, transcription and changes in shoot anatomy during Moso bamboo growth. Plant Biotechnol J 16, 72-85.
DOI URL PMID |
| [22] | Li X, Jiang DH, Yong KL, Zhang DB (2007). Varied transcriptional efficiencies of multiple Arabidopsis U6 small nuclear RNA genes. J Integr Plant Biol 49, 222-229. |
| [23] | Liang Z, Chen KL, Li TD, Zhang Y, Wang YP, Zhao Q, Liu JX, Zhang HW, Liu CM, Ran YD, Gao CX (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8, 14261. |
| [24] |
Liang Z, Zhang K, Chen KL, Gao CX (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/ Cas system. J Genet Genomics 41, 63-68.
URL PMID |
| [25] |
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274-1284.
URL PMID |
| [26] |
Marshallsay C, Connelly S, Filipowicz W (1992). Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol Biol 19, 973-983.
URL PMID |
| [27] |
Nandy S, Pathak B, Zhao S, Srivastava V (2019). Heat- shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 3, e00145.
DOI URL PMID |
| [28] |
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 691-693.
URL PMID |
| [29] |
Papikian A, Liu WL, Gallego-Bartolomé J, Jacobsen SE (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun 10, 729.
URL PMID |
| [30] |
Peng ZH, Lu Y, Li LB, Zhao Q, Feng Q, Gao ZM, Lu HY, Hu T, Yao N, Liu KY, Li Y, Fan DL, Guo YL, Li WJ, Lu YQ, Weng QJ, Zhou CC, Zhang L, Huang T, Zhao Y, Zhu CR, Liu XG, Yang XW, Wang T, Miao K, Zhuang CY, Cao XL, Tang WL, Liu GS, Liu YL, Chen J, Liu ZJ, Yuan LC, Liu ZH, Huang XH, Lu TT, Fei BH, Ning ZM, Han B, Jiang ZH (2013). The draft genome of the fast-growing non-timber forest species Moso bamboo (Phyllostachys heterocycla). Nat Genet 45, 456-461.
DOI URL PMID |
| [31] |
Ren C, Guo YC, Gathunga EK, Duan W, Li SH, Liang ZC (2019). Recovery of the non-functional EGFP-assisted identification of mutants generated by CRISPR/Cas9. Plant Cell Rep 38, 1541-1549.
DOI URL PMID |
| [32] | Sandhu M, Wani SH, Jiménez VM (2018). In vitro propagation of bamboo species through axillary shoot proliferation: a review. Plant Cell Tissue Organ Cult 132, 27-53. |
| [33] |
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014). CRISPR/Cas9- mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55, 475-481.
URL PMID |
| [34] |
Wang MB, Helliwell CA, Wu LM, Waterhouse PM, Peacock WJ, Dennis ES (2008). Hairpin RNAs derived from RNA polymerase II and polymerase III promoter-directed transgenes are processed differently in plants. RNA 14, 903-913.
URL PMID |
| [35] | Ye SW, Chen G, Kohnen MV, Wang WJ, Cai CY, Ding WS, Wu C, Gu LF, Zheng YS, Ma XQ, Lin CT, Zhu Q (2020). Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid Ma bamboo (Dendrocalamus latiflorus Munro). Plant Biotechnol J doi: 10.1111/pbi.13320. |
| [36] |
Zhang JS, Zhang H, Botella JR, Zhu JK (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol 60, 369-375.
DOI URL PMID |
| [37] |
Zhang ZZ, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li WL (2019). Development of an Agrobacterium- delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J 17, 1623-1635.
URL PMID |
| [38] | Zhao HS, Gao ZM, Wang L, Wang JL, Wang SB, Fei BH, Chen CH, Shi CC, Liu XC, Zhang HL, Lou YF, Chen LF, Sun HY, Zhou XQ, Wang SN, Zhang C, Xu H, Li LC, Yang YH, Wei YL, Yang W, Gao Q, Yang HM, Zhao SC, Jiang ZH (2018). Chromosome-level reference genome and alternative splicing atlas of Moso bamboo (Phyllostachys edulis). Gigascience 7, giy115. |
| [1] | 张强, 赵振宇, 李平华. 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024, 59(6): 978-998. |
| [2] | 胡丹玲, 孙永伟. 病毒介导的植物基因组编辑技术研究进展[J]. 植物学报, 2024, 59(3): 452-462. |
| [3] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [4] | 谭禄宾, 孙传清. 四倍体野生稻快速驯化: 启动人类新农业文明[J]. 植物学报, 2021, 56(2): 134-137. |
| [5] | 谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光. 基于CRISPR编辑系统的DNA片段删除技术[J]. 植物学报, 2021, 56(1): 44-49. |
| [6] | 苏钺凯,邱镜仁,张晗,宋振巧,王建华. CRISPR/Cas9系统在植物基因组编辑中技术改进与创新的研究进展[J]. 植物学报, 2019, 54(3): 385-395. |
| [7] | 孙建飞, 翟建云, 马元丹, 傅卢成, 卜柯丽, 王柯杨, 高岩, 张汝民. 毛竹快速生长期茎秆不同节间光合色素和光合酶活性的差异[J]. 植物学报, 2018, 53(6): 773-781. |
| [8] | 商天其, 叶诺楠, 高海卿, 高洪娣, 伊力塔. 基于多元回归树的公益林群落结构解析[J]. 植物学报, 2018, 53(2): 238-249. |
| [9] | 赵睿宇, 李正才, 王斌, 葛晓改, 戴云喜, 赵志霞, 张雨洁. 毛竹林地表覆盖年限对土壤有机碳的影响[J]. 植物生态学报, 2017, 41(4): 418-429. |
| [10] | 葛晓改, 周本智, 肖文发, 王小明, 曹永慧, 叶明. 生物质炭添加对毛竹林土壤呼吸动态和温度敏感性的影响[J]. 植物生态学报, 2017, 41(11): 1177-1189. |
| [11] | 欧阳明, 杨清培, 陈昕, 杨光耀, 施建敏, 方向民. 毛竹扩张对次生常绿阔叶林物种组成、结构与多样性的影响[J]. 生物多样性, 2016, 24(6): 649-657. |
| [12] | 杨春菊, 陈永刚, 汤孟平, 施拥军, 侯建花, 孙燕飞. 不同管理模式下毛竹幼竹的生长规律[J]. 植物学报, 2016, 51(6): 774-. |
| [13] | 陈永刚, 汤孟平, 杨春菊, 马天午, 王礼. 天然毛竹林竞争空间关系分析[J]. 植物生态学报, 2015, 39(7): 726-735. |
| [14] | 潘璐, 牟溥, 白尚斌, 古牧. 毛竹林扩张对周边森林群落菌根系统的影响[J]. 植物生态学报, 2015, 39(4): 371-382. |
| [15] | 郭慧媛, 马元丹, 王丹, 左照江, 高岩, 张汝民, 王玉魁. 模拟酸雨对毛竹叶片抗氧化酶活性及释放绿叶挥发物的影响[J]. 植物生态学报, 2014, 38(8): 896-903. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||