

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 385-395.DOI: 10.11983/CBB18151 cstr: 32102.14.CBB18151
收稿日期:2018-07-02
接受日期:2018-08-09
出版日期:2019-05-01
发布日期:2019-11-24
通讯作者:
王建华
基金资助:
Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang( )
)
Received:2018-07-02
Accepted:2018-08-09
Online:2019-05-01
Published:2019-11-24
Contact:
Jianhua Wang
摘要: CRISPR/Cas9基因组编辑技术是一项对基因组进行精准修饰的技术, 可实现对靶标基因的碱基插入、缺失或DNA片段替换。随着人们对CRISPR/Cas9系统的了解逐渐加深, 其在科研、农业和医疗等领域的应用也越来越广泛。该文简要介绍了CRISPR/Cas9基因组编辑技术的发展以及工作原理, 总结了近几年对该技术进行优化与改进的研究进展, 包括基因组编辑效率的提升、基因组编辑范围的扩展、单碱基精准编辑以及多基因同时编辑、基因组编辑安全性的提升以及基因片段替换与基因靶向转录调控, 以期为深入开展这一领域的研究提供参考。
苏钺凯,邱镜仁,张晗,宋振巧,王建华. CRISPR/Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报, 2019, 54(3): 385-395.
Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang. Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing. Chinese Bulletin of Botany, 2019, 54(3): 385-395.
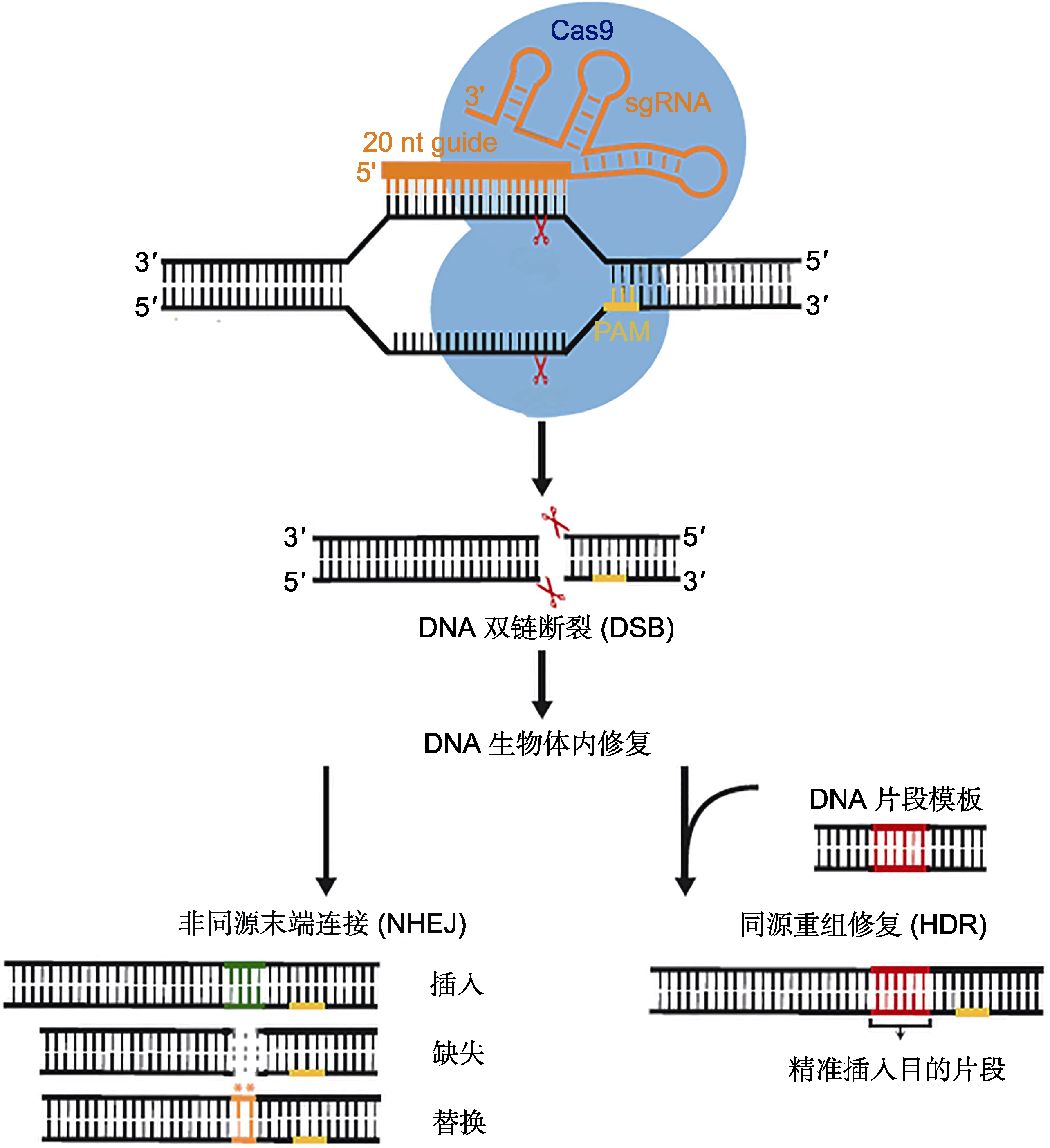
图1 CRISPR/Cas9基因组编辑原理(Jiang and Doudna, 2017)来自酿脓链球菌(Streptococcus pyogenes)的Cas9蛋白(SpCas9)与包含20 nt识别序列的向导RNA (sgRNA)结合, 然后定向切割位于原型间隔序列毗邻基序(PAM)区域上游的靶序列造成双链断裂(DSB), 最终借助生物体内自身修复实现基因组编辑。
Figure 1 The schematic diagram of CRISPR/Cas9 genome editing (Jiang and Doudna, 2017)The Cas9 protein from Streptococcus pyogenes (SpCas9) and associated guide RNA (sgRNA), containing a 20 nt recognition sequence, will cleave a target sequence located upstream of the protospacer adjacent motif (PAM) region, resulting in double-strand break (DSB). Ultimately use the repair in the organism to achieve genome editing.
| [46] | Liang Z, Chen K, Zhang Y, Liu J, Yin K, Qiu JL, Gao C ( 2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleo proteins. Nat Protoc 13, 413-430. |
| [47] |
Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh TF, Voytas DF, Zhang Y, Qi Y ( 2018). Robust transcriptional activation in plants using multiplexed CRISPR-Act 2.0 and mTALE-Act systems. Mol Plant 11, 245-256.
DOI URL |
| [48] |
Lu HP, Liu SM, Xu SL, Chen WY, Zhou X, Tan YY, Huang JZ, Shu QY ( 2017). CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol J 15, 1371-1373.
DOI URL PMID |
| [49] |
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG ( 2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274-1284.
DOI URL PMID |
| [50] | Marraffini LA, Sontheimer EJ ( 2008). CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322, 1843-1845. |
| [51] |
Meng XB, Hu XX, Liu Q, Song XG, Gao CX, Li JY, Wang KJ ( 2018). Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice. Sci China Life Sci 61, 122-125.
DOI URL |
| [52] |
Mikami M, Toki S, Endo M ( 2016). Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol 57, 1058-1068.
DOI URL PMID |
| [53] | Miki D, Zhang W, Zeng W, Feng Z, Zhu JK ( 2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9, 1967. |
| [54] |
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ ( 2011). A tale nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143-148.
DOI |
| [55] |
Mussolino C, Cathomen T ( 2013). RNA guides genome engineering. Nat Biotechnol 31, 208-209.
DOI |
| [56] | Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S ( 2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 691-693. |
| [57] |
Peng J, Wang Y, Jiang JY, Zhou XY, Song L, Wang LL, Ding C, Qin J, Liu LP, Wang WH, Liu JQ, Huang XX, Wei H, Zhang P ( 2015). Production of human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the zygotes. Sci Rep 5, 16705.
DOI URL PMID |
| [58] | Puchta H, Dujon B, Hohn B ( 1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21, 5034-5040. |
| [59] | Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F ( 2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186-191. |
| [60] |
Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, Duchateau P, Pâques F, Blanco FJ, Montoya G ( 2008). Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature 456, 107-111.
DOI URL PMID |
| [61] |
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H ( 2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant 11, 623-626.
DOI URL PMID |
| [62] |
Schiml S, Fauser F, Puchta H ( 2015). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80, 1139-1150.
DOI URL PMID |
| [63] |
Shan JW, Gao CX ( 2015). Research progress of genome editing and derivative technologies in plants. Hereditas 37, 953-973.
DOI URL PMID |
| [64] |
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C ( 2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31, 686-688.
DOI URL PMID |
| [65] | Steinert J, Schiml S, Fauser F, Puchta H ( 2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84, 1295-1305. |
| [66] |
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L ( 2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9, 628-631.
DOI URL PMID |
| [67] |
Symington LS, Gautier J ( 2011). Double-strand break end resection and repair pathway choice. Annu Rev Genet 45, 247-271.
DOI URL PMID |
| [1] |
李红, 谢卡斌 ( 2017). 植物CRISPR基因组编辑技术的新进展. 生物工程学报 33, 1700-1711.
DOI URL |
| [2] | 冉毅东, 梁振, 张毅, 高彩霞 ( 2017). 植物基因组编辑试剂材料的导入及转化系统的研究现状及前景. 中国科学: 生命科学 47, 1159-1176. |
| [68] |
Ungerer J, Pakrasi HB ( 2016). Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep 6, 39681.
DOI URL PMID |
| [69] |
Waltz E ( 2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293.
DOI URL PMID |
| [3] |
王影, 李相敢, 邱丽娟 ( 2018). CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展. 植物学报 53, 528-541.
DOI URL |
| [4] |
Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Lemaux PG, Medford JI, Orozco-Cárdenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart Jr CN ( 2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510-1520.
DOI URL PMID |
| [70] |
Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK ( 2017). Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant 10, 1007-1010.
DOI URL PMID |
| [71] |
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS ( 2015). DNA-free genome editing in plants with preassembled CRISPRCas9 ribonucleoproteins. Nat Biotechnol 33, 1162-1164.
DOI URL PMID |
| [5] |
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M ( 2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19, 1.
DOI URL |
| [6] | Baazim H ( 2014). RNA-guided transcriptional regulation in plants via dCas9 chimeric proteins. Ph.D. thesis. Thuwal, Kingdom of Saudi Arabia: King Abdullah University of Science and Technology. pp. 32-48. |
| [72] |
Xie K, Minkenberg B, Yang Y ( 2015). Boosting CRISPR/ Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112, 3570-3575.
DOI URL PMID |
| [73] |
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ ( 2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14, 327.
DOI URL PMID |
| [7] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P ( 2007). Crispr provides acquired resistance against viruses in prokaryotes. Science 315, 1709-1712.
DOI URL PMID |
| [8] |
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J ( 2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960-964.
DOI URL |
| [74] |
Xing YY, Yang J, Ren J ( 2016). Application of CRISPR/ Cas9 mediated genome editing in farm animals. Hereditas (Beijing) 38, 217-226.
DOI URL PMID |
| [75] | Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J ( 2014). Gene targeting using the Agrobacterium tumefaciens- |
| [9] | Chamberlain JR, Schwarze U, Wang PR, Hirata RK, Hankenson KD, Pace JM, Underwood RA, Song KM, Sussman M, Byers PH, Russell DW ( 2004). Gene targeting in stem cells from individuals with Osteogenesis imperfecta. Science 303, 1198-1201. |
| [10] |
Cheng AW, Jillette N, Lee P, Plaskon D, Fujiwara Y, Wang W, Taghbalout A, Wang H ( 2016). Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labelling. Cell Res 26, 254-257.
DOI URL PMID |
| [76] | mediated CRISPR-Cas system in rice.Rice 7, 5. |
| [77] |
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J ( 2017). Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15, 713-717.
DOI URL PMID |
| [78] |
Yan F, Kuang YJ, Ren B, Wang JW, Zhang DW, Lin HH, Yang B, Zhou XP, Zhou HB ( 2018). High-efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11, 631-634.
DOI URL |
| [11] |
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F ( 2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823.
DOI URL PMID |
| [12] |
Corrigan-Curay J, O’Reilly M, Kohn DB, Cannon PM, Bao G, Bushman FD, Carroll D, Cathomen T, Joung JK, Roth D, Sadelain M, Scharenberg AM, VON Kalle C, Zhang F, Jambou R, Rosenthal E, Hassani M, Singh A, Porteus MH ( 2015). Genome editing technologies: defining a path to clinic: genomic editing: establishing preclinical toxicology standards, Bethesda, Maryland 10 June 2014. Mol Ther 23, 796.
DOI URL PMID |
| [79] |
Yu Z, Chen Q, Chen W, Zhang X, Mei F, Zhang P, Zhao M, Wang X, Shi N, Jackson S, Hong Y ( 2018). Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in Arabidopsis. J Integr Plant Biol 60, 376-381.
DOI URL PMID |
| [80] |
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F ( 2015). Cpf1 is a single RNA-guided Endonuclease of a Class 2 CRISPR- Cas system. Cell 163, 759-771.
DOI URL PMID |
| [13] |
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E ( 2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602-607.
DOI URL PMID |
| [14] | Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S ( 2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390-1400. |
| [15] |
Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R ( 2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120.
DOI URL PMID |
| [16] |
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandra-segaran S ( 2005). Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33, 5978-5990.
DOI URL PMID |
| [17] |
Endo M, Mikami M, Toki S ( 2015). Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56, 41.
DOI URL PMID |
| [18] |
Feng C, Su H, Bai H, Wang R, Liu Y, Guo X, Liu C, Zhang J, Yuan J, Birchler JA, Han F ( 2018). High efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol J 16, 1848-1857.
DOI URL PMID |
| [19] |
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK ( 2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32, 279-284.
DOI URL PMID |
| [20] | Gaj T, Gersbach CA, Barbas III CF ( 2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome enginee- |
| [21] | ring.Trends Biotechnol 31, 397-405. |
| [22] |
Gasiunas G, Barrangou R, Horvath P, Siksnys V ( 2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109, E2579-E2586.
DOI URL |
| [23] |
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR ( 2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464-471.
DOI URL PMID |
| [24] |
He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y ( 2018). Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11, 1210-1213.
DOI URL PMID |
| [25] |
Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH ( 2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985-989.
DOI URL PMID |
| [26] | Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R ( 2008). Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190, 1401. |
| [27] |
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR ( 2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57-63.
DOI URL PMID |
| [28] |
Hu X, Meng X, Liu Q, Li J, Wang K ( 2017 a). Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice. Plant Biotechnol J 16, 292-297.
DOI URL PMID |
| [29] |
Hu X, Wang C, Fu Y, Liu Q, Jiao X, Wang K ( 2016). Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant 9, 943-945.
DOI URL PMID |
| [30] |
Hu X, Wang C, Liu Q, Fu Y, Wang K ( 2017 b). Targeted mutagenesis in rice using CRISPR-Cpf1 system. J Genet Genomics 44, 71-73.
DOI URL PMID |
| [31] |
Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu SM, Lee YS, Kim JS ( 2016). Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleo proteins. Nat Biotechnol 34, 807-808.
DOI URL PMID |
| [32] | Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A ( 1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169, 5429-5433. |
| [33] |
Jansen R, van Embden JD, Gaastra W, Schouls LM ( 2002). Identification of a novel family of sequence repeats among prokaryotes. OMICS 6, 23-33.
DOI URL PMID |
| [34] |
Jiang F, Doudna JA ( 2017). CRISPR-Cas9 structures and mechanisms. Ann Rev Biophy 46, 505-529.
DOI URL PMID |
| [35] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E ( 2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
DOI URL |
| [36] |
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J ( 2013). RNA-programmed genome editing in human cells. eLife 2, e00471.
DOI URL PMID |
| [37] |
Kai H, Tao X, Yuan F, Dong W, Zhu JK ( 2018). Precise A·T to G·C base editing in the rice genome. Mol Plant 11, 627-630.
DOI URL |
| [38] |
Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, Sung YH ( 2016). Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol 34, 808-810.
DOI URL PMID |
| [39] |
Kweon J, Jang AH, Kim DE, Yang JW, Yoon M, Rim SH, Kim JS, Kim Y ( 2017). Fusion guide RNAs for orthogonal gene manipulation with Cas9 and Cpf1. Nat Commun 8, 1723.
DOI URL PMID |
| [40] |
Le Blanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y ( 2017). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93, 377-386.
DOI URL PMID |
| [41] |
Li C, Zong Y, Wang YP, Jin S, Zhang DB, Song QN, Zhang R, Gao CX ( 2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19, 59.
DOI URL |
| [42] |
Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C ( 2016). Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants 2, 16139.
DOI URL PMID |
| [43] | Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J ( 2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31, 688-691. |
| [44] |
Li T, Liu B, Spalding MH, Weeks DP, Yang B ( 2012). High-efficiency talen-based gene editing produces disease-resistant rice. Nat Biotechnol 30, 390-392.
DOI URL PMID |
| [45] |
Liang PP, Xu YW, Zhang XY, Ding CH, Huang R, Zhang Z, Lv J, Xie XW, Chen YX, Li YJ, Sun Y, Bai YF, Zhou SY, Ma WB, Zhou CQ, Huang JJ ( 2015). CRISPR/Cas9- mediated gene editing in human tripronuclear zygotes. Prot Cell 6, 363-372.
DOI URL PMID |
| [1] | 王子阳, 刘升学, 杨志蕊, 秦峰. 玉米抗旱性的遗传解析[J]. 植物学报, 2024, 59(6): 883-902. |
| [2] | 张强, 赵振宇, 李平华. 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024, 59(6): 978-998. |
| [3] | 胡丹玲, 孙永伟. 病毒介导的植物基因组编辑技术研究进展[J]. 植物学报, 2024, 59(3): 452-462. |
| [4] | 苗春妍, 李铭铭, 左鑫, 丁宁, 杜家方, 李娟, 张重义, 王丰青. 湖北地黄CRISPR/Cas9基因编辑体系的建立[J]. 植物学报, 2023, 58(6): 905-916. |
| [5] | 郭彦君, 陈枫, 罗敬文, 曾为, 许文亮. 植物细胞壁木聚糖的生物合成及其应用[J]. 植物学报, 2023, 58(2): 316-334. |
| [6] | 何晓玲, 刘鹏程, 马伯军, 陈析丰. 基于CRISPR/Cas9的基因编辑技术研究进展及其在植物中的应用[J]. 植物学报, 2022, 57(4): 508-531. |
| [7] | 谭禄宾, 孙传清. 四倍体野生稻快速驯化: 启动人类新农业文明[J]. 植物学报, 2021, 56(2): 134-137. |
| [8] | 谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光. 基于CRISPR编辑系统的DNA片段删除技术[J]. 植物学报, 2021, 56(1): 44-49. |
| [9] | 凡惠金, 金康鸣, 卓仁英, 乔桂荣. 毛竹不同截短U3启动子的克隆及表达分析[J]. 植物学报, 2020, 55(3): 299-307. |
| [10] | 张璐,何新华. C3和C4植物的氮素利用机制[J]. 植物学报, 2020, 55(2): 228-239. |
| [11] | 王影, 李相敢, 邱丽娟. CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展[J]. 植物学报, 2018, 53(4): 528-541. |
| [12] | 张其生;包满珠;卢兴霞;胡惠蓉*. 大花三色堇育种研究进展[J]. 植物学报, 2010, 45(01): 128-133. |
| [13] | 周志高 汪金舫 周健民. 植物磷营养高效的分子生物学研究进展[J]. 植物学报, 2005, 22(01): 82-91. |
| [14] | 李辛雷;陈发棣. 菊花种质资源与遗传改良研究进展[J]. 植物学报, 2004, 21(04): 392-401. |
| [15] | 梁机 陈晓阳 林善枝 谢响明. 发根农杆菌Ri质粒rol基因研究进展及在林木改良上的应用[J]. 植物学报, 2002, 19(06): 650-658. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||