

植物学报 ›› 2019, Vol. 54 ›› Issue (5): 569-581.DOI: 10.11983/CBB19038 cstr: 32102.14.CBB19038
收稿日期:2019-02-26
接受日期:2019-07-09
出版日期:2019-09-01
发布日期:2020-03-10
通讯作者:
林荣呈
基金资助:
Liwen Yang,Shuangrong Liu,Rongcheng Lin( )
)
Received:2019-02-26
Accepted:2019-07-09
Online:2019-09-01
Published:2020-03-10
Contact:
Rongcheng Lin
摘要: 休眠是种子植物在长期进化过程中产生的适应性性状, 通过抑制种子在不适宜的环境中萌发进而保证植物能够在逆境中生存。此外, 休眠有助于种子的长距离运输和扩散, 因此休眠对种子延续和物种保存具有重要意义。种子由休眠向萌发的发育转变不仅关系到物种的繁衍, 而且对保证农业生产中作物的产量和品质也具有重要作用。种子的休眠和萌发受到内源激素和外源光信号的共同调控。其中, 外源光信号主要通过调控内源ABA和GA的生物合成及信号转导进而调控种子休眠和萌发。该文系统综述了外源光信号和内源激素调控种子休眠和萌发的作用通路以及两类信号通路之间的交互作用, 旨在为农业生产中利用光和激素调控种子休眠与萌发提供参考。
杨立文,刘双荣,林荣呈. 光信号与激素调控种子休眠和萌发研究进展. 植物学报, 2019, 54(5): 569-581.
Liwen Yang,Shuangrong Liu,Rongcheng Lin. Advances in Light and Hormones in Regulating Seed Dormancy and Germination. Chinese Bulletin of Botany, 2019, 54(5): 569-581.
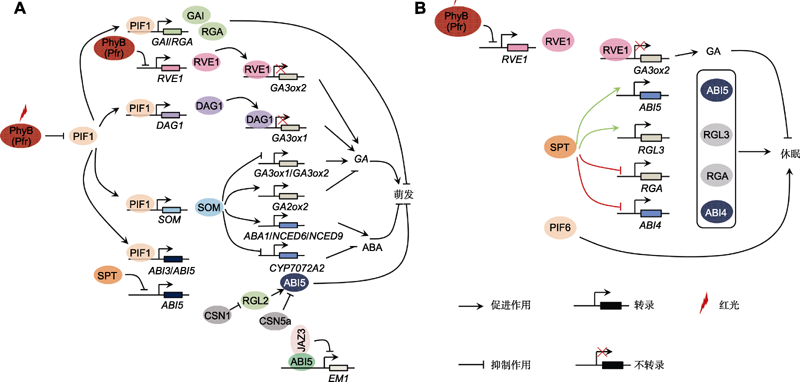
图1 光信号通过调控内源脱落酸(ABA)和赤霉素(GA)的生物合成及信号转导调控种子休眠与萌发 (A) 光信号通过调控ABA和GA通路调控种子萌发。PHYB能够介导红光促进PIF1发生泛素化降解, 从而促进种子萌发。PIF1能够通过直接激活DAG1和SOM的转录进而间接调控GA生物合成相关基因的表达, 或者直接诱导DELLA蛋白编码基因RGA和GAI的转录, 最终抑制种子萌发。同样地, PIF1也能通过调控ABA的生物合成和信号转导调控种子萌发。PIF1通过依赖于SOM的途径促进ABA生物合成, 进而抑制种子萌发; 抑或直接诱导ABI3和ABI5的转录进而促进ABA信号转导, 抑制种子萌发。除PIF1之外, PHYB还能调控RVE1的转录间接促进GA的生物合成, 最终促进种子萌发。SPT和CSN蛋白复合体通过依赖于ABI5途径调控种子萌发。SPT通过抑制ABI5的转录抑制ABA信号转导, 促进种子萌发。CSN1通过促进RGL2的泛素化降解进而抑制ABI5的蛋白稳定性, 最终促进种子萌发; 而CSN5a能够直接抑制ABI5蛋白的积累进而促进种子萌发。JAZ3通过抑制ABI5对ABA响应基因EM1的转录激活功能进而促进种子萌发。(B) 光信号通过调控ABA和GA通路调控种子休眠。PHYB能够介导红光抑制RVE1转录, 进而促进下游GA3ox2的转录, 最终抑制种子休眠。在不同生态型拟南芥背景下, SPT调控种子休眠的功能不同。其中, 在Col背景下, SPT通过促进RGL3和ABI5的转录进而促进种子休眠(绿色标识线); 在Ler背景下, SPT通过抑制RGA和ABI4的转录进而抑制种子休眠(红色标识线)。此外, PIF6也参与调控种子休眠。
Figure 1 Light signal regulates seed germination and dormancy via endogenous abscisic acid (ABA) and gibberellin (GA) biosynthesis pathway (A) Light signal regulates seed germination via ABA and GA pathway. PHYB regulates seed germination through promoting the degradation of PIF1 protein. The accumulation of PIF1 in nucleus activates the transcription of DAG1 and SOM, which indirectly regulates the expression of GA biosynthesis gene or directly induces RGA and GAI (DELLA protein encoding genes) transcription and leads to repressing seed germination. Similarly, PIF1 stimulates ABA biosynthesis and ABA signaling pathway to suppress seed germination. PIF1 induces ABA biosynthesis via SOM-dependent pathway to repress seed germination; or it induces the transcription of ABI3 and ABI5 in order to stimulate ABA signaling. Expect for PIF1, PHYB also promotes seed germination via inhibiting RVE1 transcription which indirectly promote GA biosynthesis. SPT and CSN complex could regulate seed germination in an ABI5-dependent manner. SPT suppresses ABI5 transcript to destroy ABA pathway. CSN1 stimulates RGL2 degradation to inhibit ABI5 activity, while CSN5a directly decreases the accumulation of ABI5 in order to provoke seed germination. JAZ3 promotes seed germination by repressing the transcriptional activity of ABI5 which activates ABA-responsing gene EM1 expression. (B) Light signal controls seed dormancy via ABA and GA pathway. PHYB mediates red light to repress the transcription level of GA3ox2, inhibiting seed dormancy. SPT plays different roles in regulating seed dormancy under Col and Ler background of Arabidopsis. SPT promotes seed dormancy through activating the expression of RGL3 and ABI5 under Col background (green line), however, SPT suppresses seed dormancy via inhibiting RGA and ABI4 transcript under Ler background (red line). In addition, PIF6 is also involved in controlling seed dormancy.
| 1 | 景艳军, 林荣呈 (2017). 我国植物光信号转导研究进展概述. 植物学报 52, 257-270. |
| 2 | 马朝峰, 戴思兰 (2019). 光受体介导信号转导调控植物开花研究进展. 植物学报 54, 9-22. |
| 3 | Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164, 711-729. |
| 4 | Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ (2005). Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet 111, 1365-1376. |
| 5 | Ariizumi T, Lawrence PK, Steber CM (2011). The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol 155, 765-775. |
| 6 | Bassel GW (2016). To grow or not to grow? Trends Plant Sci 21, 498-505. |
| 7 | Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103, 17042-17047. |
| 8 | Bentsink L, Koornneef M (2008). Seed dormancy and germination. Arabidopsis Book 6, e0119. |
| 9 | Bewley JD (1997). Seed germination and dormancy. Plant Cell 9, 1055-1066. |
| 10 | Bewley JD, Black M (1994). Seeds: Physiology of Development and Germination, 2nd edn. New York: Plenum Press. pp. 367. |
| 11 | Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK (1952). A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA 38, 662-666. |
| 12 | Briggs WR, Huala E (1999). Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15, 33-62. |
| 13 | Cashmore AR (2003). Cryptochromes: enabling plants and animals to determine circadian time. Cell 114, 537-543. |
| 14 | Chen F, Li BS, Li G, Charron JB, Dai MQ, Shi XR, Deng XW (2014). Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 26, 1949-1966. |
| 15 | Cheng WH, Chiang MH, Hwang SG, Lin PC (2009). Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71, 61-80. |
| 16 | Clack T, Mathews S, Sharrock RA (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25, 413-427. |
| 17 | Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H (2014). Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5, 539. |
| 18 | Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010). Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61, 651-679. |
| 19 | Davière JM, Achard P (2013). Gibberellin signaling in plants. Development 140, 1147-1151. |
| 20 | de Wit M, Galvão VC, Fankhauser C (2016). Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67, 513-537. |
| 21 | Duque P, Chua NH (2003). IMB1, a bromodomain protein induced during seed imbibition, regulates ABA- and phyA- mediated responses of germination in Arabidopsis. Plant J 35, 787-799. |
| 22 | Finch-Savage WE, Leubner-Metzger G (2006). Seed dormancy and the control of germination. New Phytol 171, 501-523. |
| 23 | Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J 70, 501-512. |
| 24 | Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J 61, 312-323. |
| 25 | Graeber K, Linkies A, Steinbrecher T, Mummenhoff K, Tarkowská D, Turečková V, Ignatz M, Sperber K, Voegele A, de Jong H, Urbanová T, Strnad M, Leubner-Metzger G (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc Natl Acad Sci USA 111, E3571-E3580. |
| 26 | Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ 35, 1769-1786. |
| 27 | Gu XY, Kianian SF, Foley ME (2006). Dormancy genes from weedy rice respond divergently to seed development environments. Genetics 172, 1199-1211. |
| 28 | Guan CM, Wang XC, Feng J, Hong SL, Liang Y, Ren B, Zuo J (2014). Cytokinin antagonizes abscisic acidmediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive5 protein in Arabidopsis. Plant Physiol 164, 1515-1526. |
| 29 | Gubler F, Millar AA, Jacobsen JV (2005). Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8, 183-187. |
| 30 | Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19, 4997-5006. |
| 31 | Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schäfer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128, 194-200. |
| 32 | Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2010). Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22, 2680-2696. |
| 33 | Holdsworth MJ, Bentsink L, Soppe WJJ (2008). Molecular networks regulating Arabidopsis seed maturation, afterripening, dormancy and germination. New Phytol 179, 33-54. |
| 34 | Hu YR, Han X, Yang ML, Zhang MH, Pan JJ, Yu DQ (2019). The transcription factor INDUCER OF CBF EXPRESSION 1 interacts with ABSCISIC ACID INSENSITIVE 5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31, 1520-1538. |
| 35 | Hu YR, Yu DQ (2014). BRASSINOSTEROID INSENSITIVE 2 interacts with ABSCISIC ACID INSENSITIVE 5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26, 4394-4408. |
| 36 | Huang X, Ouyang XH, Yang PY, Lau OS, Chen LB, Wei N, Deng XW (2013). Conversion from CUL4-based COP1- SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA 110, 16669-16674. |
| 37 | Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24, 1695-1708. |
| 38 | Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57-70. |
| 39 | Jacobsen JV, Barrero JM, Hughes T, Julkowska M, Taylor JM, Xu Q, Gubler F (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain ( Triticum aestivum L.). Planta 238, 121-138. |
| 40 | Jang IC, Yang JY, Seo HS, Chua NH (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19, 593-602. |
| 41 | Jang K, Lee HG, Jung SJ, Paek NC, Seo PJ (2015). The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci Rep 5, 12071. |
| 42 | Jiang ZM, Xu G, Jing YJ, Tang WJ, Lin RC (2016). Phytochrome B and REVEILLE1/2-mediated signaling controls seed dormancy and germination in Arabidopsis. Nat Commun 7, 12377. |
| 43 | Jin D, Wu M, Li BS, Bücker B, Keil P, Zhang SM, Li JG, Kang DM, Liu J, Dong J, Deng XW, Irish V, Wei N (2018). The COP9 signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet 14, e1007237. |
| 44 | Ju L, Jing YX, Shi PT, Liu J, Chen JS, Yan JJ, Chu JF, Chen KM, Sun JQ (2019). JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol 223, 246-260. |
| 45 | Kanai M, Nishimura M, Hayashi M (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J 62, 936-947. |
| 46 | Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light- dependent seed germination downstream of PIL5. Plant Cell 20, 1260-1277. |
| 47 | Kim W, Lee Y, Park J, Lee N, Choi G (2013). HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol 54, 555-572. |
| 48 | Lee HG, Lee K, Seo PJ (2015a). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol Biol 87, 371-381. |
| 49 | Lee K, Lee HG, Yoon S, Kim HU, Seo PJ (2015b). The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE 4 in the control of seed germination. Plant Physiol 168, 677-689. |
| 50 | Lee KP, Piskurewicz U, Turečková V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev 26, 1984-1996. |
| 51 | Lee S, Cheng H, King KE, Wang W, He YW, Hussain A, Lo J, Harberd NP, Peng JR (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAIIRGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16, 646-658. |
| 52 | Lee S, Kim SG, Park CM (2010). Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol 188, 626-637. |
| 53 | Lenser T, Theiβen G (2013). Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci 18, 704-714. |
| 54 | Li JG, Li G, Wang HY, Deng XW (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9, e0148. |
| 55 | Li XY, Chen TT, Li Y, Wang Z, Cao H, Chen FY, Li Y, Soppe WJJ, Li WL, Liu YX (2019). ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION 1 expression. Plant Cell 31, 832-847. |
| 56 | Liang T, Mei SL, Shi C, Yang Y, Peng Y, Ma LB, Wang F, Li X, Huang X, Yin YH, Liu HT (2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44, 512-523. |
| 57 | Lin CT, Shalitin D (2003). Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54, 469-496. |
| 58 | Linkies A, Müller K, Morris K, Turečková V, Wenk M, Cadman CSC, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, Leubner-Metzger G (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21, 3803-3822. |
| 59 | Liu HT, Yu XH, Li KW, Klejnot J, Yang HY, Lisiero D, Lin CT (2008a). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535-1539. |
| 60 | Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008b). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292-306. |
| 61 | Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X (2016). The NF-YC-RGL2 module integrates GA and ABA signaling to regulate seed germination in Arabidopsis. Nat Commun 7, 12768. |
| 62 | Liu X, Wang J, Yu Y, Kong LN, Liu YM, Liu ZQ, Li HY, Wei PW, Liu ML, Zhou H, Bu QY, Fang J (2019). Identification and characterization of the rice pre-harvest sprouting mutants involved in molybdenum cofactor biosynthesis. New Phytol 222, 275-285. |
| 63 | Liu XD, Zhang H, Zhao Y, Feng ZY, Li Q, Yang HQ, Luan S, Li JM, He ZH (2013a). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110, 15485-15490. |
| 64 | Liu YW, Li X, Li KW, Liu HT, Lin CT (2013b). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9, e1003861. |
| 65 | Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441-2456. |
| 66 | Ma DB, Li X, Guo YX, Chu JF, Fang S, Yan CY, Noel JP, Liu HT (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113, 224-229. |
| 67 | Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064-1068. |
| 68 | Martinez-Andújar C, Ordiz MI, Huang ZL, Nonogaki M, Beachy RN, Nonogaki H (2011). Induction of 9- cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc Natl Acad Sci USA 108, 17225-17229. |
| 69 | Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou JP, Kamiya Y, Nambara E, Truong HN (2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol 149, 949-960. |
| 70 | Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20, 55-67. |
| 71 | Née G, Kramer K, Nakabayashi K, Yuan BJ, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017b). DELAY OF GERMINATION 1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy. Nat Commun 8, 72. |
| 72 | Née G, Xiang Y, Soppe WJJ (2017a). The release of dormancy, a wake-up call for seeds to germinate. Curr Opin Plant Biol 35, 8-14. |
| 73 | Nemoto K, Ramadan A, Arimura GI, Imai K, Tomii K, Shinozaki K, Sawasaki T (2017). Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signaling by preventing GID1 degradation. Nat Commun 8, 1004. |
| 74 | Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Satoh K, Kaiwa N, Irisa T, Kinoshita T, Schroeder JI, Yates III JR, Hirayama T, Yamazaki T (2018). Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun 9, 2132. |
| 75 | Nonogaki M, Sall K, Nambara E, Nonogaki H (2014). Amplification of ABA biosynthesis and signaling through a positive feedback mechanism in seeds. Plant J 78, 527-539. |
| 76 | Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16, 3045-3058. |
| 77 | Oh E, Yamaguchi S, Hu JH, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192-1208. |
| 78 | Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47, 124-139. |
| 79 | Park J, Lee N, Kim W, Lim S, Choi G (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23, 1404-1415. |
| 80 | Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068-1071. |
| 81 | Pedmale UV, Huang SSC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, Ecker JR, Chory J (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233-245. |
| 82 | Penfield S, Hall A (2009). A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21, 1722-1732. |
| 83 | Penfield S, Josse EM, Halliday KJ (2010). A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol 73, 89-95. |
| 84 | Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995). Phytochromes: photosensory perception and signal transduction. Science 268, 675-680. |
| 85 | Ravindran P, Verma V, Stamm P, Kumar PP (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant 10, 1307-1320. |
| 86 | Ren H, Han JP, Yang PY, Mao WW, Liu X, Qiu LL, Qian CZ, Liu Y, Chen ZR, Ouyang XH, Chen X, Deng XW, Huang X (2019). Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci USA 116, 4722-4731. |
| 87 | Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Alabadí D, Masiero S (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant 8, 482-485. |
| 88 | Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, Ulm R (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103-106. |
| 89 | Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, Porri A, Sacharowski S, Gratkowska DM, Zugaj DL, Taff A, Zalewska A, Archacki R, Davis SJ, Coupland G, Koncz C, Jerzmanowski A, Sarnowski TJ (2013). DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk inArabidopsis. Plant Physiol 163, 305-317. |
| 90 | Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995-999. |
| 91 | Seo M, Nambara E, Choi G, Yamaguchi S (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69, 463-472. |
| 92 | Sharrock RA, Quail PH (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Gene Dev 3, 1745-1757. |
| 93 | Shi H, Wang X, Mo XR, Tang C, Zhong SW, Deng XW (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci USA 112, 3817-3822. |
| 94 | Shi H, Zhong SW, Mo XR, Liu N, Nezames CD, Deng XW (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25, 3770-3784. |
| 95 | Shinomura T, Nagatani A, Chory J, Furuya M (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by Phytochrome B and secondarily by Phytochrome A. Plant Physiol 104, 363-371. |
| 96 | Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93, 8129-8133. |
| 97 | Shu K, Liu XD, Xie Q, He ZH (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9, 34-45. |
| 98 | Shu K, Meng YJ, Shuai HW, Liu WG, Du JB, Liu J, Yang WY (2015). Dormancy and germination: how does the crop seed decide? Plant Biol 17, 1104-1112. |
| 99 | Shu K, Zhang HW, Wang SF, Chen ML, Wu YR, Tang SY, Liu CY, Feng YQ, Cao XF, Xie Q (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9, e1003577. |
| 100 | Silverstone AL, Ciampaglio CN, Sun TP (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155-169. |
| 101 | Simsek S, Ohm JB, Lu HY, Rugg M, Berzonsky W, Alamri MS, Mergoum M (2014). Effect of pre-harvest sprouting on physicochemical changes of proteins in wheat. J Sci Food Agric 94, 205-212. |
| 102 | Stanga JP, Smith SM, Briggs WR, Nelson DC (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163, 318-330. |
| 103 | Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y (2012). Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol 53, 107-117. |
| 104 | Vaistij FE, Barros-Galvão T, Cole AF, Gilday AD, He ZS, Li Y, Harvey D, Larson TR, Graham lA (2018). MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 115, 8442-8447. |
| 105 | Vaistij FE, Gan YB, Penfield S, Gilday AD, Dave A, He ZS, Josse EM, Choi G, Halliday KJ, Graham IA (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA 110, 10866-10871. |
| 106 | Wang YP, Li L, Ye TT, Zhao SJ, Liu Z, Feng YQ, Wu Y (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68, 249-261. |
| 107 | Wei N, Deng XW (2003). The COP9 signalosome. Annu Rev Cell Dev Biol 19, 261-286. |
| 108 | Wilson RL, Kim H, Bakshi A, Binder BM (2014). The ethylene receptors ETHYLENE RESPONSE 1 and ETHYLENE RESPONSE 2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol 165, 1353-1366. |
| 109 | Xi WY, Liu C, Hou XL, Yu H (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733-1748. |
| 110 | Xi WY, Yu H (2010). Mother of FT and TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal Behav 5, 1315-1317. |
| 111 | Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014). REDUCED DORMANCY 5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26, 4362-4375. |
| 112 | Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ (2007). Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol 64, 293-303. |
| 113 | Xu F, He SB, Zhang JY, Mao ZL, Wang WX, Li T, Hua J, Du SS, Xu PB, Li L, Lian HL, Yang HQ (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11, 523-541. |
| 114 | Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, Kamiya Y, Yamaguchi S (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol 48, 555-561. |
| 115 | Yang CW, Xie FM, Jiang YP, Li Z, Huang X, Li L (2018a). Phytochrome a negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell 44, 29-41. |
| 116 | Yang Y, Liang T, Zhang LB, Shao K, Gu XX, Shang RX, Shi N, Li X, Zhang P, Liu HT (2018b). UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4, 98-107. |
| 117 | Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun TP (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037-3057. |
| [1] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 刘旭鹏, 王敏, 韩守安, 朱学慧, 王艳蒙, 潘明启, 张雯. 植物器官脱落调控因素及分子机理研究进展[J]. 植物学报, 2025, 60(3): 472-482. |
| [3] | 孙龙, 李文博, 娄虎, 于澄, 韩宇, 胡同欣. 火干扰对兴安落叶松种子萌发的影响[J]. 植物生态学报, 2024, 48(6): 770-779. |
| [4] | 陈婷欣, 符敏, 李娜, 杨蕾蕾, 李凌飞, 钟春梅. 铁甲秋海棠DNA甲基转移酶全基因组鉴定及表达分析(长英文摘要)[J]. 植物学报, 2024, 59(5): 726-737. |
| [5] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [6] | 袁涵, 钟爱文, 刘送平, 彭焱松, 徐磊. 水毛花种子萌发特性的差异及休眠解除方法[J]. 植物生态学报, 2024, 48(5): 638-650. |
| [7] | 朱晓博, 董张, 祝梦瑾, 胡晋, 林程, 陈敏, 关亚静. 重要的种子储存物质长寿命mRNA[J]. 植物学报, 2024, 59(3): 355-372. |
| [8] | 张悦婧, 桑鹤天, 王涵琦, 石珍珍, 李丽, 王馨, 孙坤, 张继, 冯汉青. 植物对非生物胁迫系统性反应中信号传递的研究进展[J]. 植物学报, 2024, 59(1): 122-133. |
| [9] | 蔡淑钰, 刘建新, 王国夫, 吴丽元, 宋江平. 褪黑素促进镉胁迫下番茄种子萌发的调控机理[J]. 植物学报, 2023, 58(5): 720-732. |
| [10] | 唐子雯, 张冬平. 水稻胚乳淀粉积累过程的分子机理研究进展[J]. 植物学报, 2023, 58(4): 612-621. |
| [11] | 李季蔓, 靳楠, 胥毛刚, 霍举颂, 陈小云, 胡锋, 刘满强. 不同干旱水平下蚯蚓对番茄抗旱能力的影响[J]. 生物多样性, 2022, 30(7): 21488-. |
| [12] | 李聪, 齐立娟, 谷晓峰, 李继刚. 植物光信号途径重要新调控因子TZP的研究进展[J]. 植物学报, 2022, 57(5): 579-587. |
| [13] | 戴琛, 汪瑾, 卢亚萍. 衍生化UPLC-MS法测定酸性植物激素[J]. 植物学报, 2022, 57(4): 500-507. |
| [14] | 李月, 胡德升, 谭金芳, 梅浩, 王祎, 李慧, 李芳, 韩燕来. 单列毛壳菌通过促进秸秆降解并调控激素响应基因表达促进玉米生长[J]. 植物学报, 2022, 57(4): 422-433. |
| [15] | 李思源, 张照鑫, 饶良懿. 桑苗非结构性碳水化合物和生长激素对水淹胁迫的响应[J]. 植物生态学报, 2022, 46(3): 311-320. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||